5. smallanglescattering (sas)¶
This module allows smearing/desmearing of SAXS/SANS data and provides the sasImage class to read and analyse 2D detector images from SAXS cameras.
Smearing is for line collimation as in Kratky SAXS cameras and for point collimation as SAXS/SANS data. For point collimation the resolution smearing a la Pedersen is realised. This can also be used for SAXS instruments in pinhole geometry collimation with smaller pinholes and distances. Smearing as decorator of model allows simultaneous fit of smeared data from SANS/SAXS at different collimations.
Desmearing uses the Lake algorithm as an iterative procedure to desmear measured data. We follow here the improvements according to Vad using a convergence criterion and smoothing.
2D SAXS data can be read into sasImage to do typical tasks as transmission correction, masking, beamcenter location, 2D background subtraction, radial average (also for sectors).
Transmission correction and background correction are explained in Analyse SAS data
As references the waterXray scattering and a AgBe reference spectrum are available.
For form factors and structure factors see the respective modules. Conversion of sasImage to dataArray allows 2D fit of scattering patterns (see Fitting the 2D scattering of a lattice)
5.1. SAS smear/desmear 1D¶
The preferred method for convolution with instrument resolution is smear. smear modifies model functions to automatically extend the q range beyond edges for proper smearing as needed to fit smeared SANS/SAXS data. Smearing explicit data by smear or resFunct needs additional information how to extrapolate data at the detector edges. See smear for detailed explanation. Needed beamProfiles are prepared by prepareBeamProfile.
|
Smearing model/data for point collimation SANS/SAXS or line-collimated SAXS (Kratky camera) allowing simultaneous SAXS/SANS fitting. |
|
Desmearing of measured data according to Lake algorithm with possibility to stop recursion at best desmearing. |
|
Resolution smearing of small angle scattering for SANS/SAXS according to Pedersen for radial averaged data. |
|
Resolution smearing of small angle scattering for SANS/SAXS according to explict given Gaussian width in unit 1/nm. |
|
Prepare beamProfile from beam profile measurement or according to given parameters. |
|
Extract primary beam of empty cell or buffer measurement for semitransparent beam stops. |
|
Plots beam profile and weight function according to parameters in beam. |
5.2. SAS convenience¶
|
Subtract dark current, find primary beam, get transmission count and normalize by transmission and exposure time. |
|
Absolute scattering of water with components (salt, buffer) at Q=0 as reference for X-ray. |
|
The scattering intensity expected from AgBe as a reference for wavelength calibration. |
5.3. 2D sasImage¶
Read 2D image files from SAXS cameras and extract the corresponding data.
The sasImage is a 2D array that allows direct subtraction and multiplication (e.g. transmission) respecting given masks in operations. E.g.
sample=js.sas.sasImage('sample.tiff')
solvent=js.sas.sasImage('solvent.tiff')
corrected = sample/sampletransmission - solvent/solventtransmission
Image manipulation like Gaussian filter from scipy.ndimage can be used.
Calibration of detector distance including offset detector positions, radial average, size reduction and more. .pickCenter allows sensitive detection of the beamcenter in SAS geometry. .calibrateOffsetDetector allows sensitive calibration of the detector position parameters.
TIFF images are read directly. Other image formats as .edf can be treated using the library fabio. Example is given in
sasImage.
Examples are shown in sasImage including reading of different
images produced by 2D X-ray detectors using the fabio library.
|
Creates/reads sasImage as maskedArray from a detector image or array. |
5.3.1. sasImage methods¶
- Proccessing/Calibration
asImage([scale, colormap, inverse, ...])Returns the sasImage as 8bit RGB image using PIL.
saveAsTIF(filename[, fill])Save the sasImage as float32 tif without loosing information.
radialAverage([center, number, kind, ...])Radial average of image and conversion to wavevector q.
azimuthAverage([center, qrange, number, ...])Azimuthal average of image and conversion to wavevector q.
recalibrateDetDistance([center, number, ...])Recalibration of detectorDistance by AgBe reference for point collimation.
calibrateOffsetDetector([lattice, center, ...])Compare sasImage to calibration standard in powder average to determine detector position.
gaussianFilter([sigma])Gaussian filter in place.
getPolar([center, scaleR, offset])Transform to polar coordinates around center with interpolation.
showPolar([center, scaleR, offset, scale])Show image transformed to polar coordinates around center.
reduceSize([bin, center, border])Reduce size of image using uniform average in box.
show(**kwargs)Show sasImage as matplotlib figure.
Strip of all attributes and return a simple array without mask.
asdataArray([masked])Return representation of sasImage as dataArray with (qx, qy, qz, I(qx,qy,qz)).
interpolateMaskedRadial([radial])Interpolate masked values from radial averaged image or function.
pickBeamcenter([levels, symmetry])Pick the beam center from a calibration sample as AgBe in standard SAS geometry if a beamstop is used.
findCenterOfIntensity([center, size])Find beam center as center of intensity..
- line collimation feature
lineFindCenter([minmax, use, show, darkline])Find center of primary beam for line collimation cameras with semitransparent beamstop.
lineAverage([sigma, darkline, whiteline])Line average and conversion to wavevector q for Kratky (line) collimation cameras.
- Masking
Reset the mask.
maskFromImage(image)Use/copy mask from image.
maskRegion(xmin, xmax, ymin, ymax)Mask rectangular region.
maskRegions(regions)Mask several regions.
maskbelowLine(p1, p2)Mask points at one side of line.
maskTriangle(p1, p2, p3[, invert])Mask inside triangle.
mask4Polygon(p1, p2, p3, p4[, invert])Mask inside polygon of 4 points.
maskCircle(center, radius[, invert])Mask points inside circle.
maskSectors(angles, width[, radialmax, invert])Mask sector around center.
- Attributes
3D scattering vector \(q\) for pixels with detector placed in standard SAS or offset geometry.
3D scattering vector \(|q|\) for detector pixel.
pQaxes()Get scattering vector along detector pixel axes X, Y around center.
Show specific attribute names as sorted list of attribute names.
showattr([maxlength, exclude])Show specific attributes with values as overview.
setAttrFromImage(image)Copy center, detector_distance, alpha, beta, gamma wavelength, pixel_size from image.
setDetectorPosition(center, detector_distance)Set parameters describing the position and orientation of the detector.
setDetectorDistance(detector_distance)Set detector distance as shortest distance to sample along plane normal vector.
setPlaneCenter(center)Set beamcenter or center of detector plane where plane normal has shortest distance to sample.
setPlaneOrientation([alpha, beta, gamma])Set orientation angles of detector plane .
setPixelSize(pixel_size)Set pixel_size.
setWavelength(wavelength)Set wavelength.
getfromcomment(name[, replace, newname])Extract name from .artist or .imageDescription with attribute name in front.
5.4. 2D sasImage convenience¶
|
Create .png files from grayscale images with log scale conversion to values between [1,255]. |
|
Create text file with image descriptions as list of content. |
|
Read a list of images returning sasImage`s. |
5.5. Housekeeping¶
|
Opens and reads a SAXS data file in the .pdh (Primary Data Handling) format. |
|
Scales elements of data to have same mean .Y value in the overlap region of .X . |
|
Takes a dataset and removes single spikes from data by substitution with spline. |
|
Takes a dataset and removes single spikes. |
|
Locate all files matching supplied filename pattern in and below supplied root directory. |
|
Copies all files matching pattern in tree below root to destination directory |
|
Adds the parameters stored in xml part of a .pdh file as eg. |
|
Read SAXSPACE .pdh files and removes spikes by removeSpikes. |
This module allows smearing/desmearing of SAXS/SANS data and provides the sasImage class to read and analyse 2D detector images from SAXS cameras.
Smearing is for line collimation as in Kratky SAXS cameras and for point collimation as SAXS/SANS data. For point collimation the resolution smearing a la Pedersen is realised. This can also be used for SAXS instruments in pinhole geometry collimation with smaller pinholes and distances. Smearing as decorator of model allows simultaneous fit of smeared data from SANS/SAXS at different collimations.
Desmearing uses the Lake algorithm as an iterative procedure to desmear measured data. We follow here the improvements according to Vad using a convergence criterion and smoothing.
2D SAXS data can be read into sasImage to do typical tasks as transmission correction, masking, beamcenter location, 2D background subtraction, radial average (also for sectors).
Transmission correction and background correction are explained in Analyse SAS data
As references the waterXray scattering and a AgBe reference spectrum are available.
For form factors and structure factors see the respective modules. Conversion of sasImage to dataArray allows 2D fit of scattering patterns (see Fitting the 2D scattering of a lattice)
- jscatter.smallanglescattering.AgBeReference(q, wavelength, n=array([1, 2, 3, 4, 5, 6, 7, 8, 9]), ampn=None, domainsize=100, udw=0.1, asym=0, lg=1)[source]¶
The scattering intensity expected from AgBe as a reference for wavelength calibration.
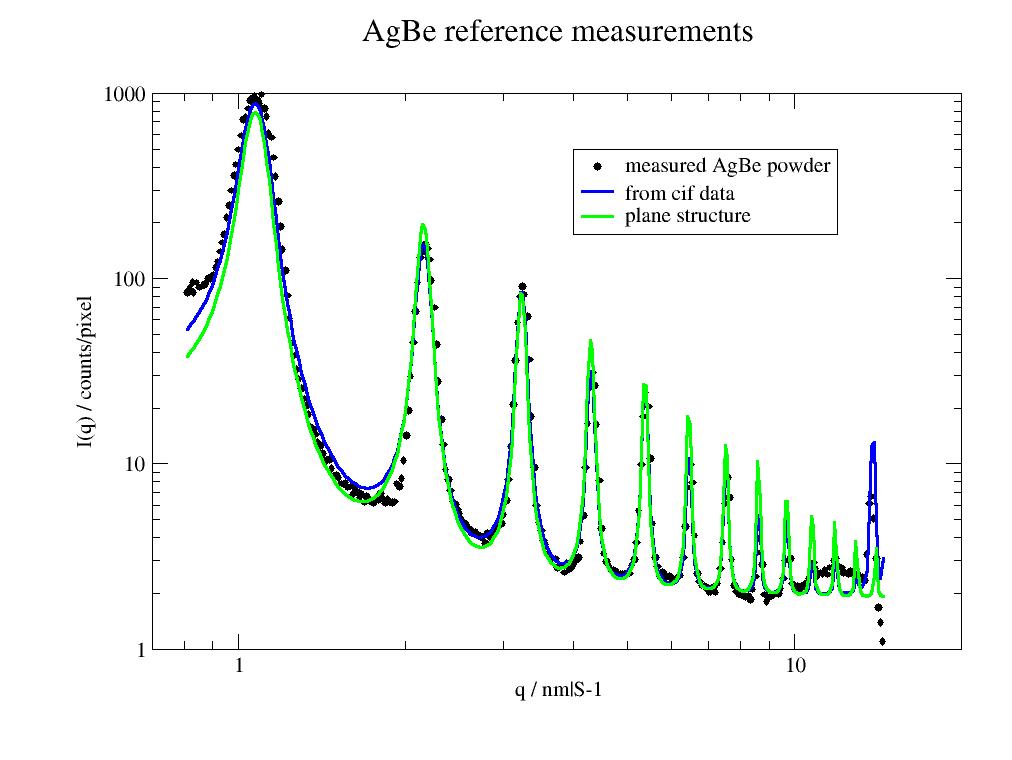
The intensities assume a d-spacing of 5.8378 nm and a reduction of the intensity as q**-2. The domain size determines the width according to Scherrer equation [2]. The first peak is at 1.076 1/nm. The result needs to be convoluted with the instrument resolution by resFunct or smear. Here only the main planes of AgBe are taken into account. See example.
- Parameters:
- qarray
Wavevector in units 1/nm.
- wavelengthfloat
Wavelength, units nm.
- narray of int
Order of the peaks to calculate.
- ampnlist of float
Amplitudes of the peaks.
- domainsizefloat
Domainsize of AgBe crystals in nm. default 100 nm as is given in [1].
- udwfloat
Displacement u in Debye Waller factor exp(-u**2*q**2/3) in units nm.
- asymfloat
Factor asymmetry in Voigt function describing the peaks.
- lgfloat
Lorenzian/gaussian fraction of both FWHM, describes the contributions of gaussian and lorenzian shape. See Voigt for details.
- Returns:
- dataArray
References
[1]T. C. Huang, H. Toraya, T. N. Blanton and Y. Wu X-ray Powder Diffraction Analysis of Silver Behenate, a Possible Low-Angle Diffraction Standard J. Appl.Cryst.(1993).26,180-184
[2]Patterson, A. The Scherrer Formula for X-Ray Particle Size Determination Phys. Rev. 56 (10): 978–982 (1939) doi:10.1103/PhysRev.56.978.
Examples
Xray AgBe measurement
The simplified plane model is good enough for peak determination while the crystallographic model gets the high order peaks intensities better.
import numpy as np import jscatter as js # # Look at raw calibration measurement calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') bc=calibration.center calibration.mask4Polygon([bc[0]+8,bc[1]],[bc[0]-8,bc[1]],[bc[0]-8+60,0],[bc[0]+8+60,0]) # mask center calibration.maskCircle(calibration.center, 18) # mask outside shadow calibration.maskCircle([500,320], 280,invert=True) # calibration.show(axis='pixel',scale='log') cal=calibration.radialAverage() # lattice from crystallographic data in cif file. agbe=js.sf.latticeFromCIF(js.examples.datapath + '/1507774.cif',size=[0,0,0]) sfagbe=js.sf.latticeStructureFactor(cal.X, lattice=agbe, domainsize=50, rmsd=0.001, lg=1, hklmax=17,wavelength=0.15406) # simplified model of planes ag = js.sas.AgBeReference(q=sfagbe.X, wavelength=0.13414, n=np.r_[1:14], domainsize=50) p=js.grace() p.plot(cal, le='measured AgBe powder') # add scaling and background (because of unscaled raw data) p.plot(sfagbe.X,190*sfagbe.Y+1.9,sy=0,li=[1,3,4],legend='from cif data') p.plot(ag.X, ag.Y*3+1.9, sy=0, li=[1,3,5],le='plane structure') p.yaxis(scale='log',label='I(q) / counts/pixel') p.xaxis(scale='log',label='q / nm|S-1',min=0.7,max=20) p.title('AgBe reference measurements') p.legend(x=4,y=500) # p.save(js.examples.imagepath+'/AgBeplanes.jpg')
- jscatter.smallanglescattering.addXMLParameter(data)[source]¶
Adds the parameters stored in xml part of a .pdh file as eg. in SAXSPACE .pdh files.
- Parameters:
- datadataArray
Already read pdh file. XML content is found in comments of the read files and starts with ‘<’.
- jscatter.smallanglescattering.autoscaleYinoverlapX(dataa, key=None, keep='lowest')[source]¶
Scales elements of data to have same mean .Y value in the overlap region of .X .
- Parameters:
- dataadataList
Data to scale
- keystring
Data are grouped into unique values of attribute key before scaling. E.g. to do it for a series of concentrations for each concentration individually.
- keepdefault ‘l’
If ‘l’ the lowest X are kept and higher X are scaled successively to next lower X. Anything else highest X are kept and other are scaled to next higher.
- Returns:
- dataList
new scaled dataList
Notes
First data are sorted along .X.mean() scaling value is stored in .autoscalefactor
- jscatter.smallanglescattering.copyFiles(pattern, root='.', destination='copy', link=False)[source]¶
Copies all files matching pattern in tree below root to destination directory
- Parameters:
- patternfile pattern
Pattern used in fnmatch.filter
- rootdirectory, default is os.curdir
Directory where to start
- destinationdirname
Destination
- linkbool
If True links are created.
- jscatter.smallanglescattering.desmear(Ios, beamProfile, NIterations=-25, windowsize=4, qmax=4, output=True, **kwargs)[source]¶
Desmearing of measured data according to Lake algorithm with possibility to stop recursion at best desmearing.
For negative NIterations the iterations are stopped if a convergence criterion reaches a minimum as described by Vad [2]. In each step a smoothing based on the ratio desmeared/observed as described in [2] is used (point average with windowsize).
- Parameters:
- IosdataArray
Original smeared data
- beamProfiledataArray
Beam profile as prepared with prepareBeamProfile
- NIterationsint, default=-15 (<0 automatic mode)
Number of iterations to stop. automatic mode: NIterations<0: stops when convergence criterion increases again (as described by Vad [2]) and abs(NIterations) is maximum number of iterations.
- qmaxfloat, default=4
Maximum in scattering vector q up to where the convergence criterion is evaluated. This reduces the influence of the noise at larger a.
- windowsizeodd int , default=4
Window size for smoothing in each step of desmearing (running average).
- outputbool
Print output.
- Returns:
- dataArray
Notes
The Lake algorithm approximates the unsmeared scattering cross section iteratively (n>0, with \(u_0(q)=I_{observed}(q)\)) as
\[u_n(q) = \frac{I_{observed}(q)u_{n-1}(q)}{\int_{q`}u_{n-1}(q`)R(q,q`)dq`}\]The iterative method converges if [2]
\[\gamma_n(q) = \frac{I_{observed}(q)}{\int_{q`}u_{n-1}(q`)R(q,q`)dq`} \to 1\]The convergence criterion is
\[|<\gamma_n(q)>-1| = minimum\]which means that the iterative algorithm is stopped if \(<\gamma_n(q)>\) increases again because amplified noise leads to increasing \(<\gamma_n(q)>\).
Here \(<>\) is the average over all \(q\) and \(R(...)\) is the resolution function smearing the measurement.
References
[1]Lake, J. A. (1967). Acta Cryst. 23, 191–194.
Examples
- jscatter.smallanglescattering.getBeamWidth(empty, minmax='auto', show=False)[source]¶
Extract primary beam of empty cell or buffer measurement for semitransparent beam stops. Used for Kratky camera.
The primary beam is searched and cut between the next minima found, then normalized. Additionally a Gaussian fit is done and hwhm is included in result profile.
- Parameters:
- emptydataArray
Empty cell measurement with the transmitted beam included.
- minmax‘auto’,[float,float]
Automatic or interval for search of primary beam. E.g. [-0.03,0.03] allow for explicitly setting the interval.
- showbool
Show the fit result
- Returns:
- dataArray with beam width profile
.sigma sigma of fit with Gaussian .hwhm half width half maximum
- jscatter.smallanglescattering.locateFiles(pattern, root='.')[source]¶
Locate all files matching supplied filename pattern in and below supplied root directory.
- Parameters:
- patternfile pattern
Pattern used in fnmatch.filter
- rootdirectory, default is os.curdir
Directory where to start
- Returns:
- File list
- jscatter.smallanglescattering.moveSAXSPACE(pattern, root='./', destination='./despiked', medwindow=5, SGwindow=5, sigma=0.2, order=2)[source]¶
Read SAXSPACE .pdh files and removes spikes by removeSpikes.
This is mainly for use at JCNS SAXSPACE with CCD camera as detector :-))))
- Parameters:
- patternstring
Search pattern for filenames
- rootstring
Root path
- destinationstring
Where to save the files
- medwindowodd integer
Window size of scipy.signal.medfilt
- SGwindowodd int, None
Savitzky-Golay filter see scipy.signal.savgol_filter
- orderint
Polynominal order of scipy.signal.savgol_filter
- sigmafloat
Deviation factor of eY If datapoint-median> sigma*std its a spike
Notes
Default values are adjusted to typical SAXSPACE measurement.
- jscatter.smallanglescattering.plotBeamProfile(beam, p=None)[source]¶
Plots beam profile and weight function according to parameters in beam. Used for Kratky camera.
- Parameters:
- beam
beam with parameters
- pGracePlot instance
Reuse the given plot
- jscatter.smallanglescattering.prepareBeamProfile(data=None, **kwargs)[source]¶
Prepare beamProfile from beam profile measurement or according to given parameters.
- Parameters:
- datadataArray,’trapez’,’SANS’, float
- Contains measured beam profile, explicit Gaussian width list or type ‘SANS’, ‘trapz’.
dataArray: Beam profile measurement for line collimation as measured during adjusting of a Kratky camera. The beam profile looks like a trapez. The measured beam profile will be smoothed and made symmetric.
dataArray + keyword ‘explicit’: Explicit given **Gaussian width for each Q* value (see below). Missing values will be interpolated. The explicit given width should have same units as scattering vector q in the data. Needs additional parameter explicit set. E.g. all data from KWS2@MLZ have this in 4th column of data.
float : A beam profile (similar to ‘explicit’) with a single Gaussian width will be used. The explicit given width should have same units as scattering vector q in the data. This can be used for explicit measured beam profile from the attenuated primary beam in SAXS/SANS if it looks Gaussian.
- ‘trapez’Line collimation with trapezoidal parameters
Needs: a, b, bxw, dIW.
- ‘SANS’Smearing a la Pedersen; see resFunct for parameters
Needs: collDist, collAperture, detDist, sampleAperture, wavelength, wavespread, dpixelWidth, dringwidth, extrapolfunc
This can also be used for SAXS with appropriate values for wavespread and aperture sizes.
- collDist,collAperture,detDist,sampleAperturefloat
Parameters as described in resFunct for SANS. These are determined from the experimental setup.
- wavelength,wavespread,dpixelWidth,dringwidth,extrapolfuncfloat
Parameters as described in resFunct for SANS. These are determined from the experimental setup.
- explicitint
For explicit given Gaussian width the index of the column with the width. For merged dataFiles of KWS2@MLZ this is the forth column with index 3. The width should have same units as q.
- afloat
Larger full length of trapezoidal profile in detector q units. Ignored for measured profile.
- bfloat
Shorter full length of trapezoidal profile in detector q units If a=b ==> a=a*(1+1e-7), b=b*(1-1e-7) Ignored for measured profile.
- bxwfloat,dataArray
Beam width profile. Use getBeamWidth to cut the primary beam and fit a Gaussian. A float describes the beam half-width at half maximum (hwhm of Gaussian). If bxw is the profile prepared by getBeamWidth the experimental profile is used.
- dIWfloat
Detector slit width in detector q units. Length on detector to integrate parallel to beam length for line collimation. On my SAXSpace this is 1.332 as given in the header of the file (20 mm in real coordinates).
- wavelengthfloat,
Wavelength in nm default 0.155418 nm for SAXS 0.5 nm for SANS
- detDistfloat, default 305.3558
Detector distance in units mm Default is detector distance of SAXSpace
- Returns:
- beamProfile as dataArray
Notes
For measured beam profiles in line collimation parameters a,b are determined from the flanks for trapezoidal profile.
Detector q units are equivalent to the pixel distance as expressed in a corrected measurement.
For ‘explicit’ Gaussian width a SANS measurement as on KWS2 can be used which has sigma as 4th column. Missing values are interpolated.
Be careful with merged data sets. If merged data sets with different resolutions are used a part of the q values might be smeared with wrong resolution width. It is meanwhile a common BAD practice. At least there should be no overlap in q range showing consecutive q with alternating resolution. The option ‘explicit’ should work correctly if for all q values the corresponding beamProfile contains a respective width. If these are missing the interpolation of missing values will fail and alternate between next neighbour resolution. The best practice is to use non merged data sets.
Examples
# use as # a single width for all e.g. primary beam measurement fbeam= js.sas.prepareBeamProfile(0.01) # prepare measured beamprofile for line collimation mbeam = js.sas.prepareBeamProfile(beam, bxw=0.01, dIW=1.) # prepare profile with trapezoidal shape (a,b are fitted above) tbeam = js.sas.prepareBeamProfile('trapz', a=mbeam.a, b=mbeam.b, bxw=0.01, dIW=1) # prepare profile SANS (missing parameters get defaults) Sbeam = js.sas.prepareBeamProfile('SANS', detDist=2000,wavelength=0.4,wavespread=0.15) # prepare profile with explicit given Gaussian width in column 3 as e.g. KWS2@MLZ Gbeam = js.sas.prepareBeamProfile(measurement,explicit=3)
- jscatter.smallanglescattering.readpdh(pdhFileName)[source]¶
Opens and reads a SAXS data file in the .pdh (Primary Data Handling) format.
If data contain X values <0 it is assumed that the primary beam is included as for SAXSpace instruments. Use js.sas.transmissionCorrection to subtract dark counts and do transmission correction.
- Parameters:
- pdhFileNamestring
File name
- Returns:
- dataArray
Notes
Alternatively the files can be read ignoring the information in the header (it is stored in the comments)
data=dA(pdhFileName,lines2parameter=[2,3,4])
PDH format used in the PCG SAXS software suite developed by the Glatter group at the University of Graz, Austria. This format is described in the appendix of the PCG manual (below from version 4.05.12 page 159).
In the PDH format, lines 1-5 contain header information, followed by the SAXS data.
line 1: format A80 -> description line 2: format 16(A4,1X) -> description in 16x4 character groups (1X = space separated) line 3: format 8(I9,1X) -> 8 integers (1X = space separated) line 4: format 5(E14.6,1X) -> 5 float (1X = space separated) line 5: format 5(E14.6,1X -> 5 float(1X = space separated) line 6+ format 3(E14.6,1X) - SAXS data x,y,error (1X = space separated)
- with:
line 3 field 0 : number of points
line 4 field 4 : normalization constant (default 1, never zero!!)
Anything else can have different meanings.
- The SAXSpace and SAXSess (AntonPaar) format add:
line 4 field 2 : detector distance in mm
line 4 field 5 : wavelength
line 5 field 2 : detector slit length (equivalent to width of integration area) in q units
Additional xml parameter as in the SAXSPACE format appended can be extracted to attributes by addXMLParameter. Mainly this is “Exposure” time.
Example data for SAXSpace
<emptyline> SAXS BOX 2057 0 0 0 0 0 0 0 0.000000E+00 3.052516E+02 0.000000E+00 1.000000E+00 1.541800E-01 0.000000E+00 1.332843E+00 0.000000E+00 0.000000E+00 0.000000E+00 -1.033335E-01 2.241656E+03 1.024389E+00 -1.001430E-01 2.199972E+03 1.052537E+00 ...
- jscatter.smallanglescattering.removeSpikes(dataa, xmin=None, xmax=None, medwindow=5, SGwindow=None, sigma=0.2, SGorder=2)[source]¶
Takes a dataset and removes single spikes.
A median filter is used to find single spikes. If abs(data.Y-medianY)/data.eY>sigma then the medianY value is used. If SGwindow!=None Savitzky-Golay filtered values are used. If sigma is 0 then new values (median or Savitzky-Golay filtered) are used everywhere.
- Parameters:
- xmin,xmaxfloat
Minimum and maximum X values
- dataadataArray
Dataset with eY data
- medwindowodd integer
window size of scipy.signal.medfilt
- SGwindowodd int, None
Savitzky-Golay filter see scipy.signal.savgol_filter without the spikes; window should be smaller than instrument resolution
- SGorderint
Polynomial order of scipy.signal.savgol_filter needs to be smaller than SGwindow
- sigmafloat
Relative deviation from eY If datapoint-median> sigma*eY its a spike
- Returns:
- Filtered and smoothed dataArray
- jscatter.smallanglescattering.removeSpikesMinmaxMethod(dataa, order=7, sigma=2, nrepeat=1, removePoints=None)[source]¶
Takes a dataset and removes single spikes from data by substitution with spline.
Find minima and maxima of data including double point spikes; no 3 point spikes scipy.signal.argrelextrema with np.greater and np.less are used to find extrema
- Parameters:
- dataadataArray
Dataset with eY data.
- orderint
Number of points see scipy.signal.argrelextrema. Distance between extrema.
- sigmafloat
Deviation factor from std dev; from eY. If datapoint -spline> sigma*std its a spike.
- nrepeatint
Repeat the procedure nrepeat times.
- removePointslist of integer
Instrument related points to remove because of dead pixels. ‘JCNS’ results in a list for SAXSPACE at JCNS Jülich.
- Returns:
- data with spikes removed
- jscatter.smallanglescattering.resFunct(S, collDist=8000.0, collAperture=10, detDist=8000.0, sampleAperture=10, wavelength=0.5, wavespread=0.2, dpixelWidth=10, dringwidth=1, extrapolfunc=None)[source]¶
Resolution smearing of small angle scattering for SANS/SAXS according to Pedersen for radial averaged data.
\(I(q_0)= \int R(q,q_0)S(q)dq\) with Kernel \(R(q,q0)\) of equ. 33 in [1] including wavelength spread, finite collimation and detector resolution. Default parameters are typical for a SANS machine like KWS2@JCNS with rectangular apertures.
To smear model functions (also for fitting) please use smear directly, which handles extrapolation at edges automatically.
If explicit data are smeared (not a model) edges can be extrapolated as power law, Guinier like or constant. Best practice is to calculate the used model for extended Q range, smear it and prune to the needed data range. This is demonstrated in example 2.
- Parameters:
- Sarray like
Model scattering function as dataArray with X. and .Y q in units 1/nm .Y can be arbitrary unit.
- collDistfloat, default 8000
Collimation distance in mm. If 0 unsmeared data are returned.
- collAperturefloat, default 10
Collimation aperture rectangular size in mm
- detDistfloat, default 8000
Detector distance in mm. If 0 unsmeared data are returned.
- sampleAperturefloat, default 10
Sample aperture rectangular size in mm
- wavelengthfloat, default 0.5
Wavelength in nm
- wavespreadfloat, default 0.1
FWHM wavelengthspread dlambda/lambda
- dpixelWidthfloat, default 10
Detector pixel width in mm
- dringwidthinteger, default 1
Number of pixel for averaging
- extrapolfunclist , default None
Type of extrapolation at low and high X edges as list for [low edge,high edge]. If a singe value is given this is used for both.
‘’ : no interpolation
Low edge towards zero
float : Power law extrapolation
None : A constant value as Y(X.min()).
anything else : Low X data are log scaled, then X**2 extrapolated as Guinier like extrapolation.
High edge towards infinity
float : Power law extrapolation of low X e.g. a=-4 for q**a for Porod scaling.
None : A constant value as Y(X.max()).
- Returns:
- dataArray [‘wavevector; smeared scattering; unsmeared scattering; half width smearing function’]
Notes
HalfWidthSmearingFunction is the FWHM the Gaussian used for smearing including all effects.
The resolution is assumed to be equal in direction parallel and perpendicular to q on a 2D detector as described in chap. 2.5 in [1].
We neglect additional smearing due to radial averaging (last paragraph in chap 2.5 of [1]).
Defaults correspond to a typical medium resolution measurement.
extrapolfunc allows extrapolation at both edges to reduce edge effects. The best values depends on the measured signal shape at the edge. The optimal way is to calculate the used model for the whole Q range, smear it and prune to the needed range. This is demonstrated in example 2. If at low q only a power law is observed use the corresponding extrapolation. Same for high q.
An example for SANS fitting with resFunc is given in example_SANSsmearing.py.
References
Examples
Reproducing Table 1 of [1]
import jscatter as js q=js.loglist(0.1,10,500) S=js.ff.sphere(q,6) # this is the direct call of resFunc, use smear instead as shown in next example Sr=js.sas.resFunct(S, collDist=2000.0, collAperture=20, detDist=2000.0, sampleAperture=10, wavelength=0.5, wavespread=0.2,dpixelWidth=0,dringwidth=0) # plot it p=js.grace() p.plot( S,sy=[1,0.3],li=1,legend='sphere') p.plot( Sr,sy=[2,0.3,2],li=2,legend='smeared sphere') p.plot(Sr.X,Sr[-1],li=4,sy=0,legend='FWHM in nm\S-1 ') p.yaxis(min=1e-3,scale='l',charsize=1.5,label='I(q) / a.u.',tick=[10,9]) p.yaxis(min=1e-1,tick=[10,9]) p.xaxis(scale='l',charsize=1.5,label='q / nm\S-1',tick=[10,9]) p.legend(x=0.8,y=5e5)
Example 2:
Smear model over full range and interpolate to needed data. This is a way to smear a model for fitting, but is not possible for desmearing of measured data. smear extends the q range according to the width and does proper integration without interpolation. smear the preferred method.
meas=js.dA('measureddata.dat') # load data # define profile resol2m = js.sas.prepareBeamProfile('SANS', detDist=2000,collDist=2000.,wavelength=0.4,wavespread=0.15) q=np.r_[0.01:5:0.01] # or extend q range as np.r_[0:meas.X.min():0.01,meas.X,meas.X.max():meas.X.max()*2:0.1] # calc model temp=js.ff.ellipsoid(q,2,3) # smear it smearedmodel=js.sas.smear(temp,resol2m).interpolate(X=meas.X)
- jscatter.smallanglescattering.resFunctExplicit(S, beamprofile, extrapolfunc=None)[source]¶
Resolution smearing of small angle scattering for SANS/SAXS according to explict given Gaussian width in unit 1/nm. Used in smear.
\(I(q_0)= \int R(q,q_0)S(q)dq\) with Gaussian kernel \(R(q,q0)\) in equ. 33 in [1]_. E.g. for merged dataFiles of KWS2@MLZ the explicit width is given in the 4th column.
To smear model functions (also for fitting) please use smear directly, which handles extrapolation at edges automatically.
- Parameters:
- Sarray like
Scattering function (model) as dataArray with X. and .Y q in units 1/nm, .Y can be arbitrary unit
- beamprofilebeamProfile prepared by ‘explicit’ or float
Beam profile as prepared from prepareBeamProfile with ‘explicit’ given values for all q or with a single width for all q. See prepareBeamProfile.
- extrapolfunclist , default None
Type of extrapolation at low and high X edges for handling of the border as list for [low edge,high edge]. If a singe value is given this is used for both.
‘’ : no interpolation
Low edge
float : Power law extrapolation of low X e.g. a=-4 for q**a for Porod scaling.
None : A constant value as Y(X.min()).
anything else : Low X data are log scaled, then X**2 extrapolated as Guinier like extrapolation.
High edge
float : Power law extrapolation of low X e.g. a=-4 for q**a for Porod scaling.
None : A constant value as Y(X.max()).
- Returns:
- dataArray
columns [‘wavevector; smeared scattering; unsmeared scattering; half width smearing function’]
Notes
HalfWidthSmearingFunction is the FWHM the Gaussian used for smearing including all effects.
- jscatter.smallanglescattering.smear(unsmeared=None, *, beamProfile=None, **smkwargs)[source]¶
Smearing model/data for point collimation SANS/SAXS or line-collimated SAXS (Kratky camera) allowing simultaneous SAXS/SANS fitting.
The beamProfile parameter has to be given as keyword argument
beamProfile=mybeam.- Smearing at detector edges involves evaluation of the model at extrapolated q values.
For model functions smear is used as decorator
@smearand the q range is automatically extended.Simultaneous fitting of SAXS/SANS (inclusive multiple detector distances) the data need corresponding
.beamProfileas attribute. During fitting.beamprofileis used for smearing of the model. See Notes for usage.For explicit given data extrapolation at each edge needs to be chosen according to the observed behaviour at the edge (eg power law at low q or constant at high q). See prepareBeamProfile for the options.
Call only with keyword arguments. See Notes for details.
- Parameters:
- unsmearedfunction or dataArray
Model function or explicit data to be smeared. The scattering vector in the model function should have name starting with one of ‘qQ’. If used as a decorator the function is inserted automatically by the @smear syntax and should not be given. If any smearing related parameter is None as detDist=None smearing is bypassed. The unit of Q must be 1/nm
- beamProfilebeamProfile or ‘trap’, ‘SANS’, ‘explicit’, dataArray
Beamprofile that determines smearing as prepared from prepareBeamProfile. Always use as keyword argument If no beamProfile is given input and smkwargs are used in prepareBeamProfile to define beamprofile. Measured Profile is also treated by prepareBeamProfile. For explicit profile the width unit must be 1/nm.
- smkwargs
See prepareBeamProfile for kwargs.
- Returns:
- dataArray
for line collimation: smeared data
- other: dataArray with columns
[q, smeared data, original data, half width smearing function] and additional attributes as defined in the respective beamProfiles and smeared model.
Notes
Smearing a model function as decorator (like @smear ) for fitting.
Decorators modify functions. Here the model is smeared according to the provided beamprofile extending the q range at edges.
The model scattering vector must have a name starting with one of “qQ”. Units for Q is 1/nm.
Beamprofile parameters as detDist are updated for smearing during the function call. This allows that during fitting also beamprofile parameters can be fitted. Setting any of the beamprofile parameters to None bypasses smearing (e.g. for SAXS or simulation).
Use as :
# define resolution resol2m = js.sas.prepareBeamProfile('SANS', detDist=2000,collDist=2000.,wavelength=0.4,wavespread=0.10, collAperture=10, sampleAperture=6, dpixelWidth=10, dringwidth=1) # decorate model to automatically smear it (dont use `unsmeared` argument as it is added automatically) @js.sas.smear(beamProfile=resol2m) def smearedSphere(q,R,bgr,detDist=3000,collDist=2000.,c=1): sp = js.ff.sphere(q=q,radius=R,contrast=c) sp.Y=sp.Y+bgr return sp # for fitting with attribute detDist and collDist in fitted data for different detector distances sp=smearedSphere(q=q,R=13,detDist=8000,collDist=8000.)
The above decorator is equivalent to the following. Use smearedSphere as smeared model function.
def sphere(q,R,bgr,detDist=3000,collDist=2000.,contrast=1): sp = js.ff.sphere(q,R,contrast) sp.Y=sp.Y+bgr return sp smearedsphere=js.sas.smear(sphere,beamProfile=resol2m)
To fit SAXS and SANS data together (also with changing beamprofile parameters as collimation) use the parameter beamProfile in the model. If calling the smeared model with a valid beamProfile this is used instead of the one defined in @smear One may remind that also pinhole SAXS has a smearing even if it is small. Ask your instrument responsible for reasonable values or do a measurement of the primary beam.
This allows fitting with different profiles if the corresponding data contain .beamProfile as attribute. See Fitting multiple smeared data together.
fbeam = js.sas.prepareBeamProfile(0.02) fbeam2 = js.sas.prepareBeamProfile(0.01) # define smeared model with beamProfile as parameter @js.sas.smear(beamProfile=fbeam) def sphere(q,R,bgr,contrast=1, beamProfile=None): sp = js.ff.sphere(q=q,radius=R,contrast=contrast) sp.Y=sp.Y+bgr return sp # call as sphere(q=q,R=13,bgr=0,beamProfile=fbeam2) # to fit data add corresponding beamProfile attribute to each dataArray in a dataList dll[0].beamProfile=fbeam dll[1].beamProfile=fbeam2 dll.fit(sphere,.....)
Using merged data (multiple detector distances in one set of data with overlap) is possible with ‘explicit’ beamProfile if (in particular in the overlap region):
no q value is used twice
for each value a resolution width exists
The resolution width is given in same units as q.
Only this guarantees that always the correct width is used (depending on detector distance). Usually this is fulfilled if you prepare the beamProfile with data containing the width as last column like merged SANS data.
Smear explicit data
Explicit data show artifacts due to smearing at the edges if extrapolation is done wrong. The default is a constant value from the edge which is good for a low q plateau or a high q flat background. For the above model smearing this is not important as the q range is automatically extended. For desmearing e.g. of multi detector distance SANS data this is important if detector edges are within a non constant region.
import jscatter as js #define beamprofile resol8m = js.sas.prepareBeamProfile('SANS', detDist=8000,collDist=8000.,wavelength=0.4,wavespread=0.10, collAperture=15, sampleAperture=5, dpixelWidth=10, dringwidth=1,) resol8m2 = js.sas.prepareBeamProfile('SANS', detDist=8000,collDist=8000.,wavelength=0.4,wavespread=0.10, collAperture=15, sampleAperture=5, dpixelWidth=10, dringwidth=1, extrapolfunc=['guinier',None]) # create some data q=js.loglist(0.01,1,300) sp=js.ff.sphere(q,radius=13) # smear data spsmeared=js.sas.smear(sp,beamProfile=resol8m) # smear in a limited range, default is extrapolation with constant edge value spsmeared2=js.sas.smear(sp[:,sp.X>0.2],beamProfile=resol8m) # guinier like extrapolation spsmeared2ex=js.sas.smear(sp[:,sp.X>0.2],beamProfile=resol8m2) # plot it p=js.grace(1,1) p.plot(sp) p.plot(spsmeared,li=[1,3,1],sy=0) p.plot(spsmeared2,li=[1,3,2],sy=0,le='low edge underestimate') p.plot(spsmeared2ex,li=[1,3,3],sy=0,le='low edge improved') p.yaxis(scale='log',min=10000,max=1e8) p.legend(x=0.2,y=1e5) p.new_graph(xmin=0.5,xmax=0.9,ymin=0.55,ymax=0.9) xs=(spsmeared.X>0.18) & (spsmeared.X<0.25) p[1].plot(spsmeared[:,xs],li=[1,3,1],sy=0) xs=(spsmeared2.X>0.18) & (spsmeared2.X<0.25) p[1].plot(spsmeared2[:,xs],li=[1,3,2],sy=0) p[1].plot(spsmeared2ex[:,xs],li=[1,3,3],sy=0) p[1].yaxis(scale='log',min=8e6,max=2e7) p[0].title('Examine smearing at edge')
If unsmeared has attributes a, b, dIW, bxw, detDist these are used for the beamProfile, if not given in function call.
If wavelength is missing in data a default of 0.155418 nm for Xray \(K_{\alpha}\) line is assumed. For SANS 0.5 nm.
During smearing for Kratky camera an integration over the beam width and beam length are performed. In this integration \(q_{w,l}= ((q+q_{w})^2)+q_{l}^2)^{1/2}\) is used with \(q_{w}\) along the beam width and \(q_{l}\) along the beam length. in regions \(q_{w,l} > max(q_{data})\) we estimate the measured scattering intensity by the mean of the last 10 points of the measured spectra to allow for a maximum in \(q\) range. The strictly valid q range can be estimated by calculating \(q_{x,y} < max(q)\) with 2 times the used beam width and beam length. As the smearing for larger \(q\) has no real effect the estimate might be still ok.
Examples
For more examples see How to build a more complex model, How to fit SANS data including the resolution for different detector distances and Smearing and desmearing of SAX and SANS data.
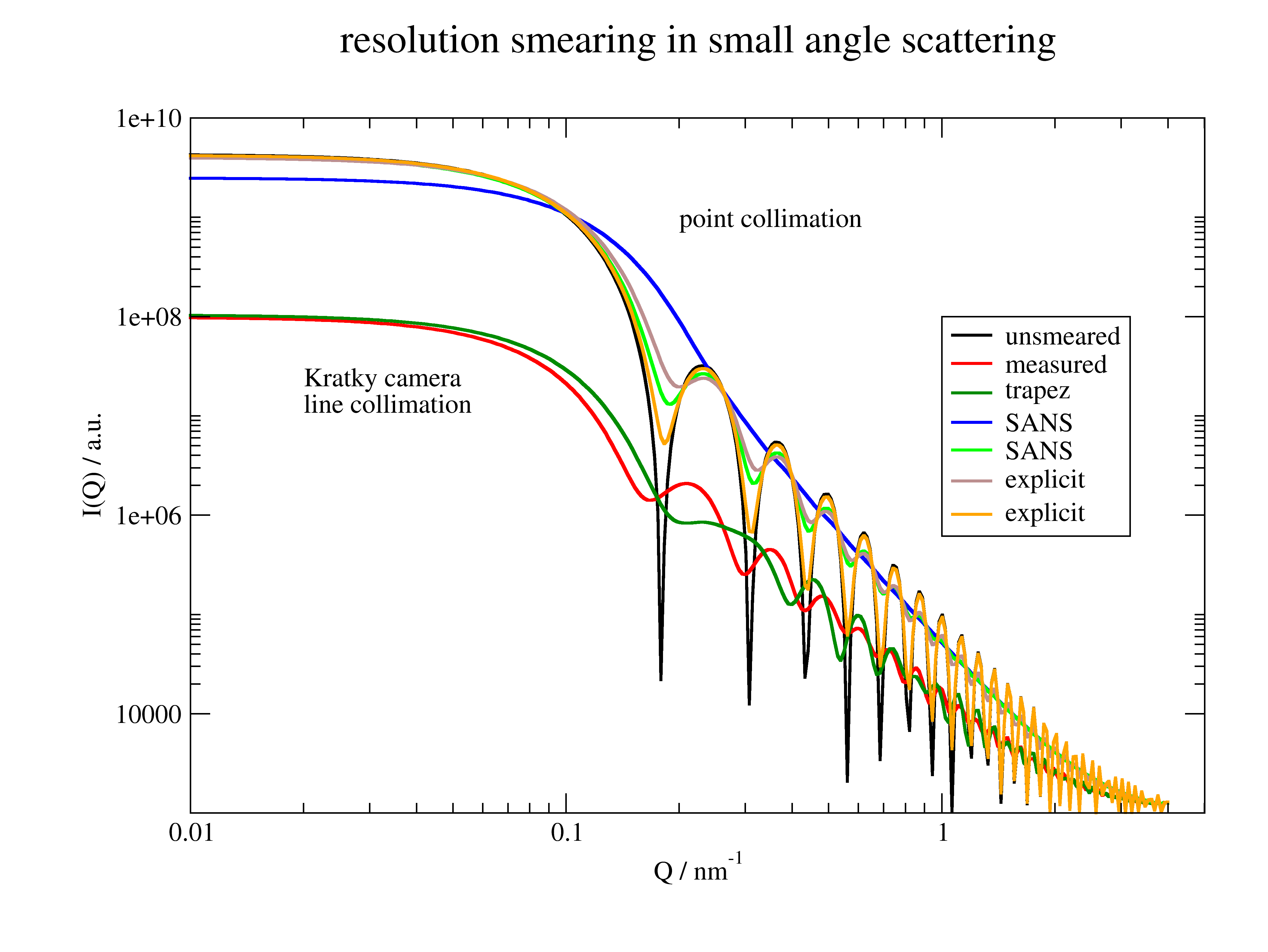
import jscatter as js q=js.loglist(0.01,4,300) def sphere(q,R,bgr,contrast=1): sp = js.ff.sphere(q,R,contrast) sp.Y=sp.Y+bgr return sp # # prepare different beamprofiles # measured Kratky camera BeamProfile.pdh for line collimation beam = js.sas.readpdh(js.examples.datapath+'/BeamProfile.pdh') mbeam = js.sas.prepareBeamProfile(beam , bxw=0.01, dIW=1.) # # Kratky camera with explicit given beamprofile parameters describing trapezoidal beam profile tbeam = js.sas.prepareBeamProfile('trapez', a=mbeam.a, b=mbeam.b, bxw=0.01, dIW=1) # # different SANS detector distances, see prepareBeamProfile for default parameters Sbeam1 = js.sas.prepareBeamProfile('SANS', detDist=2000,wavelength=0.4,wavespread=0.1,sampleAperture=5) Sbeam2 = js.sas.prepareBeamProfile('SANS', detDist=20000,wavelength=0.4,wavespread=0.1,sampleAperture=5) # # Using an explicit given resolution width in column 3 # here q and resolution width (column 3) must have units 1/nm i70=js.dL(js.examples.datapath+'/polymer.dat')[0] Gbeam = js.sas.prepareBeamProfile(i70,explicit=3) # # Using a single width e.g. for point collimation SAXS data # the width corresponds to Gaussian width from attenuated empty beam measurement fbeam = js.sas.prepareBeamProfile(0.01) # smear data data = sphere(q,R=25,bgr=1000) datasm = js.sas.smear(unsmeared=data,beamProfile=mbeam) datast = js.sas.smear(unsmeared=data,beamProfile=tbeam) datasS1 = js.sas.smear(unsmeared=data,beamProfile=Sbeam1) datasS2 = js.sas.smear(unsmeared=data,beamProfile=Sbeam2) datasG = js.sas.smear(unsmeared=data,beamProfile=Gbeam) datasf = js.sas.smear(unsmeared=data,beamProfile=fbeam) p=js.grace() p.title('resolution smearing in small angle scattering') p.plot(data,li=[1,2,-1],sy=0,le='unsmeared') for dat in [datasm,datast,datasS1,datasS2,datasG,datasf]: p.plot(dat,li=[1,2,-1],sy=0,le=dat.beamProfile.beamProfType) p.yaxis(scale='l') p.xaxis(scale='l') p.legend(x=1,y=1e8) p.text(r'Kratky camera \nline collimation' , x=0.02,y=2e7) p.text('point collimation' , x=0.2,y=8e8) p.xaxis(label=r'Q / nm\S-1') p.yaxis(label=r'I(Q) / a.u.') #p.save(js.examples.imagepath+'/smearingexamples.png')
- jscatter.smallanglescattering.transmissionCorrection(data, dark=None, emptybeam=None, edge=0.03, exposure=None, transmission=None, Izero=None)[source]¶
Subtract dark current, find primary beam, get transmission count and normalize by transmission and exposure time. IN PLACE.
For measurements including the primary beam (no beamstop or from semitransparent beamstop) or with explicit given parameters. The maximum of the primary beam peak after dark subtraction determines transmission counts for the first.
- Parameters:
- datadataArray
- A raw measurement
From a SAXSpace instrument read by js.dA(‘filename.pdh’,lines2parameter=[2,3,4]) including primary beam.
From a SAXSpace measurement read as sasImage and averaged using image.lineAverage including primary beam and background subtracted (use dark = None).
Ganesha radial averaged data with additional parameters.
For other data the missing parameters need to be given explicitly.
- darkdataArray, None
- Dark current measurement \(I_{dark}\).
None,0: Dark is already subtracted.
dataArray: A dark measurements including dead pixel. (these are corrected doing the subtraction).
float: Dark as counts/pixel/s. E.g. a value <0.1/3600s for Dectris detectors as noted in the datasheet.
- emptybeamdataArray, float, default=None
- Empty beam measurement \(I_b\) to subtract from measurement.
If it is a raw measurement (not transmission corrected) first transmissionCorrection is applied. emptybeam.transmission==1 and .primarypeakmean is given in units (counts/s).
If float (in units counts/s) this value is used as emptybeam.primarypeakmean for calculation of the actual sample transmission.
- edgefloat, default 0.03
Wavevector value below beam stop edge. Also the primary beam is searched below this value.
- exposureoptional, float, default None
Exposure time \(t_{exposure}\) in unit ‘s’. If not given explicit it is extracted from the data :
For SAXSpace data the xml description at the end of the file is examined.
For Ganesha data from
.exposure_time[0]
- transmissionoptional, float, str, None
- Transmission T of the data measurement.
float: explicitly given transmission T.
‘edge’: detect as average below edge.
If primary beam is included (SAXSpace) from the peak height.
From Ganesha data from entry
.transmission_factor[0]
- Izerofloat, None
- Intensity primary beam \(I_0\) for normalisation if different detector distances are used.
For Ganesha measurements taken from
.Izero[0](units counts/s).Ignored for SAXSpace Izero=1.
- Returns:
- None, IN PLACE
Adds attributes .centerTransmissionPeak, .centerTransmissionPeakMax and .transmission as relative transmission to primary beam for samples for SAXSSpace.
If emptybeam is NOT given the transmission value is the maximum peak value in counts/s. The same is for evaluating the emptybeam measurement itself. To compare the treated emptybeam multiply with this maximum peak value.
Notes
- After using
transmissionCorrectionthe data contain (also in given emptybeam) \(I' = \big(\frac{I-I_{dark}}{T}-I_{b}T\big) /I_0/t_{exposure}\)
Correction
Brulet at al [1] describe the data correction for SANS, which is in principle also valid for SAXS, if incoherent contributions are neglected.
The difference is, that SAXS has typical transmission around ~0.3 for 1mm water sample in quartz cell due to absorption, while in SANS typical values are around ~0.9 for D2O. Larger volume fractions in the sample play a more important rule for SANS as hydrogenated ingredients reduce the transmission significantly, while in SAXS still the water and the cell (quartz) dominate.
One finds for a sample inside of a container with thicknesses (\(z\)) for sample, buffer (solvent), empty cell and empty beam measurement (omitting the overall q dependence):
\[I_s = \frac{1}{z_S}\big((\frac{I_S-I_{dark}}{T_S}-I_{b}T_S\big) - \big(\frac{I_{EC}-I_{dark}}{T_{EC}}-I_{b}T_{EC})\big) - \frac{1}{z_B}\big((\frac{I_B-I_{dark}}{T_B}-I_{b}T_B\big) - \big(\frac{I_{EC}-I_{dark}}{T_{EC}}-I_{b}T_{EC})\big)\]- where
\(I_s\) is the interesting species
\(I_S\) is the sample of species in solvent (buffer)
\(I_B\) is the pure solvent (describing a constant background)
\(I_{dark}\) is the dark current measurement
\(I_b\) is the empty beam measurement
\(I_{EC}\) is the empty cell measurement
\(z_x\) corresponding sample thickness
\(T_x\) corresponding transmission as primary beam intensity ratio \(T_x=I_x(0)/I_b(0)\)
The recurring pattern \(\big((\frac{I-I_{dark}}{T}-I_{b}T\big)\) shows that the primary beam (not absorbed by the beam stop) is attenuated by the corresponding sample contributing unscattered intensity.
For equal sample thickness \(z\) the empty beam is included in subtraction of \(I_B\):
\[I_s = \frac{1}{z} \big((\frac{I_S-I_{dark}}{T_S}-I_{b}T_S) - (\frac{I_B-I_{dark}}{T_B}-I_{b}T_B)\big)\]The simple case
If the transmissions are nearly equal as for e.g. protein samples with low concentration (\(T_S \approx T_B\)) we only need to subtract the transmission and dark current corrected buffer measurement from the sample.
\[I_s = \frac{1}{z} \big((\frac{I_S-I_{dark}}{T_S}) - (\frac{I_B-I_{dark}}{T_B}\big)\]Higher accuracy for large volume fractions
For larger volume fractions \(\Phi\) the transmission might be different and we have to take into account that only \(1-\Phi\) of solvent contributes to \(I_S\). We may incorporate this in the sense of an optical density changing the effective thickness \(\frac{1}{z_B}\rightarrow\frac{1-\Phi}{z_B}\) resulting in different thicknesses \(z_S \neq z_B\)
Transmission
The transmission is measured as the ratio \(T=\frac{I(q=0)_{sample}}{I(q=0)_{emptybeam}}\) with \(I(q=0)\) as the primary beam intensity.
If the primary beam tail is neglected in the above equation \(I(q=0)_{emptybeam}\) only gives a common scaling factor and can be omitted if arbitrary units are used. Alternatively one can scale to the EC transmission with \(T_{EC}=1\) For absolute calibration the same needs to be done. One finds \(T_{sample in cell}=T_{empty cell}T_{sample without cell}\).
References
[1]Improvement of data treatment in small-angle neutron scattering Brûlet et al.Journal of Applied Crystallography 40, 165-177 (2007)
- jscatter.smallanglescattering.waterXrayScattering(composition='h2o1', T=293, units='mol')[source]¶
Absolute scattering of water with components (salt, buffer) at Q=0 as reference for X-ray.
According to [1] a buffer of water with components might be used. Ions need to be given separatly as [‘55.51h2o1’,’0.15Na1’,’0.15Cl1’] for 0.15 M NaCl solution. It is accounted for the temperature dependence of water density and compressibility.
- Parameters:
- compositionstring
Buffer composition as in scatteringLengthDensityCalc give dissociated ions separatly as [‘1Na’,’1Cl’] with concentration in mol prepended the additional scattering as ionic liquid of the ions in water is taken into account see [1] . mass in g; 1000g water are 55.508 mol
- Tfloat
Temperature in °K
- units‘mol’
Anything except ‘mol’ prepended unit is mass fraction. ‘mol’ prepended units is mol and mass fraction is calculated as \(mass=[mol] mass_{molecule}\) e.g. 1l Water with 123 mmol NaCl [‘55.508H2O1’,’0.123Na1’,’0.123Cl1’]
- Returns:
- float: absolute scattering length in Units 1/cm.
Notes
\(I(0)=(\sigma_{water}^2f_e^2 n_{ew}^2 k_B T \chi + \sum_{ci} n_i N_A 1000 n_{ei}^2 f_e^2 )/100\)
with
\(\sigma_{water}\) water density
\(\chi\) compressibility
\(n_{ew}\) number of electrons per water molecule
\(f_e\) cross section of electron in nm
\(k_B\) Boltzmann constant
\(n_i\) concentration component i
\(n_{ei}\) number of electrons per molecule component i in Mol
\(\sum_{ci}\) is done for all ions separately if given
For references regarding
waterdensity()andwatercompressibility()see respective functions in formel.References
[1] (1,2)A high sensitivity pinhole camera for soft condensed matter T. Zemb, O. Tache, F. Né, and O. Spalla, J. Appl. Crystallogr. 36, 800 (2003).
[2]SAXS experiments on absolute scale with Kratky systems using water as a secondary standard Doris Orthaber et al. J. Appl. Cryst. (2000). 33, 218±225
Examples
# pure H2O js.sas.waterXrayScattering(['55.5h2o1']) # 0.0164 1/cm # phosphate buffer js.sas.waterXrayScattering(['55.5h2o1','0.2na2h1P1O4','0.2na1H1p1o4','0.1h1']) # 0.0965 1/cm # 1M NaCl buffer js.sas.waterXrayScattering(['55.5h2o1','1na1','1cl1']) # 0.0360 1/cm # 100mM NaCl buffer js.sas.waterXrayScattering(['55.5h2o1','0.1na1','0.1cl1']) # 0.0183 1/cm
Read 2D image files from SAXS cameras and extract the corresponding data.
The sasImage is a 2D array that allows direct subtraction and multiplication (e.g. transmission) respecting given masks in operations. E.g.
sample=js.sas.sasImage('sample.tiff')
solvent=js.sas.sasImage('solvent.tiff')
corrected = sample/sampletransmission - solvent/solventtransmission
Image manipulation like Gaussian filter from scipy.ndimage can be used.
Calibration of detector distance including offset detector positions, radial average, size reduction and more. .pickCenter allows sensitive detection of the beamcenter in SAS geometry. .calibrateOffsetDetector allows sensitive calibration of the detector position parameters.
TIFF images are read directly. Other image formats as .edf can be treated using the library fabio. Example is given in
sasImage.
Examples are shown in sasImage including reading of different
images produced by 2D X-ray detectors using the fabio library.
- jscatter.sasimagelib.AgBepeaks = [1.0753, 2.1521, 3.2286, 4.3049, 5.3813, 6.4576, 7.5339, 8.6102, 9.6865, 10.7628]¶
AgBe peak positions
- class jscatter.sasimagelib.PickerBeamCenter(circle, image, destination, symmetry=6)[source]¶
Bases:
objectClass to pick the center of calibration sasImage
- circle :
Circle around center to move
- image :
image to use for calculation of profiles
- destination :
axes where to plot profiles
- symmetry :
number of sectors to use for averaging
- class jscatter.sasimagelib.PickerDetPosition(image, lattice, axes, buttons, latticeparameters)[source]¶
Bases:
objectClass to pick the detector position of calibration sasImage
We have 2 axes with the image and the lines to align. We use buttons to update the image parameters directly and use a single update method for all.
- class jscatter.sasimagelib.SubArray(arr)[source]¶
Bases:
ndarraySubclass used in sasImage.
don’t use this directly as intended use is through sasImage.
Defines a generic np.ndarray subclass, that stores some metadata in attributes It seems to be the default way for subclassing maskedArrays to have the array_finalize from this subclass.
- property array¶
As bare array
- property attr¶
Show specific attribute names as sorted list of attribute names.
- setattr(objekt, prepend='', keyadd='_')[source]¶
Set (copy) attributes from objekt.
- Parameters:
- objektobjekt with attr or dictionary
Can be a dictionary of names:value pairs like {‘name’:[1,2,3,7,9]} If object has property attr the returned attributenames are copied.
- prependstring, default ‘’
Prepend this string to all attribute names.
- keyaddchar, default=’_’
If reserved attributes (T, mean, ..) are found the name is ‘T’+keyadd
- jscatter.sasimagelib.createImageDescriptions(images)[source]¶
Create text file with image descriptions as list of content.
- Parameters:
- imageslist of sasImages or glob pattern
List of images
- Returns:
- file
Examples
import jscatter as js images = js.sas.readImages('*.tiff') js.sas.createImageDescriptions(images)
- jscatter.sasimagelib.createImageFromArray(data, xgrid=None, zgrid=None, method='nearest', fill_value=0)[source]¶
Create sasImage from 2D dataArray with .X and .Z coordinates and .Y values.
If points are missing these are interpolated using .regrid.
- Parameters:
- datadataArray
Data to create image.
- xgridarray, None, int
New grid in x dimension. If None the unique values in .X are used. For integer the xgrid with these number of points between [min(X),max(X)] is generated.
- zgrid :array, None
New grid in z dimension (second dimension). If None the unique values in .Z are used. For integer the zgrid with these number of points between [min(X),max(X)] is generated.
- methodfloat,’linear’, ‘nearest’, ‘cubic’
Filling value for new points as float or order of interpolation between existing points. See griddata
- fill_value
Value used to fill in for requested points outside of the convex hull of the input points. See griddata
- Returns:
- sasImage
Examples
import jscatter as js import numpy as np import matplotlib.pyplot as pyplot import matplotlib.tri as tri def func(x, y): return x*(1-x)*np.cos(4*np.pi*x) * np.sin(4*np.pi*y**2)**2 # create random points in [0,1] xz = np.random.rand(1000, 2) v = func(xz[:,0], xz[:,1]) # create dataArray data=js.dA(np.stack([xz[:,0], xz[:,1],v],axis=0),XYeYeX=[0, 2, None, None, 1, None]) newdata=data.regrid(np.r_[0:1:100j],np.r_[0:1:200j],method='cubic') newdata.Y+=newdata.Y.max() image=js.sas.createImageFromArray(newdata,100,100) image.show()
- jscatter.sasimagelib.createLogPNG(filenames, center=None, size=None, colormap='jet', equalize=False, contrast=None)[source]¶
Create .png files from grayscale images with log scale conversion to values between [1,255].
This generates images viewable in simple image viewers as overview. The new files are stored in the same folder as the original files.
- Parameters:
- filenamesstring
Filename with glob pattern as ‘file*.tif’
- center[int,int]
Center of crop region.
- sizeint
- Size of crop region.
If center is given a box with 2*size around center is used.
If center is None the border is cut by size.
- colormapstring, None
Colormap from matplotlib or None for grayscale. For standard colormap names look in mpl.showColors().
- equalizebool
Equalize the images.
- contrastNone, float
Autocontrast for the image. The value (0.1=10%) determines how much percent are cut from the intensity histogram before linear spread of intensities.
- jscatter.sasimagelib.readImages(filenames, onlyheaders=False)[source]¶
Read a list of images returning sasImage`s.
- Parameters:
- filenamesstring
Glob pattern to read
- Returns:
- list of sasImage`s
Notes
To get a list of image descriptions:
images=js.sas.readImages(path+'/latest*.tiff') [i.description for i in images]
- jscatter.sasimagelib.sImemoize(cachesize=4, attrnames=[])[source]¶
A least-recently-used cache decorator to cache attributes from sasImages dependent on the position attributes.
Cache is according to attribute names of the used class given as list in attrnames. default is empty list
We use this to avoid multiple calculation of some sasImage parameters for same detector settings Therefore a small cachesize is ok.
- class jscatter.sasimagelib.sasImage(file, detector_distance=None, center=None, alpha=None, beta=None, gamma=None, pixel_size=None, wavelength=None, copy=None, maskbelow=0, flip=None, onlyheaders=False)[source]¶
Bases:
SubArray,MaskedArrayCreates/reads sasImage as maskedArray from a detector image or array.
All methods of maskedArrays including masking of invalid areas work.
Masked areas are automatically masked for all math operations.
Arithmetic operations for sasImages work as for numpy arrays e.g. to subtract background image or multiplying with transmission or corrections [1]. Use the numpy.ma methods.
Pixel coordinates in images are [height,width] with origin located at upper-left corner.
- Parameters:
- filestring, PIL.Image, ndarray
- Filename to read as image or created PIL.Image/ndarray to process.
TIFF Images (‘.tiff’) are read and information in the EXIF tag is used if present.
EDF, Fit2dImage, TIF (‘.tif’) and more are read (using internally fabio [2] ). The according metadata are converted to attributes. See second example.
A list of readable file formats is found at https://github.com/silx-kit/fabio
numpy arrays with ndim=2 can be used directly
A PIL.Image can be read with .open or created directly from an numpy array, see Notes.
If not present add manually metadata as detector_distance, center, pixelsize using the sasImage methods (see examples). All information found in the files is accessible as attributes. To get an overview with values use .showattr().
- detector_distancefloat, sasImage, optional
Detector distance from calibration measurement or calibrated image in unit m. Overwrites value in the file EXIF tag.
- centerNone, list 2xfloat, sasImage, optional
Center is [height, width] pixel position of closest point to sample or primary beam in standard SAS geometry with origin located at upper-left corner. Overwrites value given in the file EXIF tag.
- alphafloat, optional
Rotation angle between incident beam and detector plane normal in degree.
- betafloat, optional
Rotation angle of Detector pixel X dimension around detector plane normal in degree.
- gammafloat, optional
Rotation of detector normal around incident beam in degree.
- pixel_size[float,float], optional
Pixel size in [x,y] direction in units m.
- wavelengthfloat, optional
Wavelength in units angstrom.
- copysasImage, optional
Copy center, detector_distance, alpha, beta, gamma, wavelength, pixel_size from sasImage. Overwrites corresponding data from file.
- maskbelowfloat, default =0, optional
Mask values below this value.
- flipint, default None
Reverse the order of pixels for an axis (0=vertical, 1=horizontal). Other data are not modified, only pixel data are flipped.
- Returns:
- imagesasImage with attributes
.center : center
.iX : Height pixel positions
.iY : Width pixel positions
.filename
.artist : Additional attributes from EXIF Tag Artist
.imageDescription : Additional attributes from EXIF Tag ImageDescription
.detector_distance
.alpha, .beta, .gamma as detector plane orientation
.wavelength wavelength in units Å.
.pixel_size in units m.
Notes
Unmasked data can be accessed as .data
The mask is .mask and initial set to all negative values.
Masking of a pixel is done as
image[i,j]=np.ma.masked. Use mask methods as implemented.Geometry mask methods can be used and additional masking methods from numpy masked Arrays.
import jscatter as js from numpy import ma cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') cal.mask = ma.nomask # reset mask cal[cal<0]= ma.masked # mask negative values cal[(cal>30) & (cal<100)] = ma.masked # mask region of values
TIFF tags with index above 700 are ignored.
Tested for reading tiff image files from Pilatus detectors as given from our metal jet SAXS machines Ganesha and Galaxi at JCNS, Jülich.
Tested with .edf images from ALBA and from ID02 beamline at ESRF.
Additional SAXSpace TIFF files are supported which show frames per pixel on the Y axis. This allows to examine the time evolution of the measurement on these line collimation cameras (Kratky camera). Instead of the old PIL the newer fork Pillow is needed for the multi page TIFFs. Additional the pixel_size is set to 0.024 (µm) as for the JCNS CCD camera.
Center & orientation:
The x,y orientation for images are not well defined and dependent on the implementation on the specific camera setup. Typically coordinates are used in [height,width] with the origin in the upper left corner. This is opposed to the expectation of [x,y] coordinates with the X horizontal and the origin at the lower-left. To depict 2D images in the way we expect it from the experimental setup (location of the center, orientation) it is not useful to change orientation. Correspondingly the first coordinate (usually expected X) is the height coordinate in vertical direction.
For convenient reading of several images:
Read calibration measurement as
cal=js.sas.sasImage('mycalibration.tif')
Determine detector distance and center (both saved in cal as attribute).
cal.pickBeamcenter() cal.recalibrateDetDistance()
Read following sasImages copying the information stored in sasImage
calbysample=js.sas.sasImage('nextsample.tif', copy=cal)
- A PIL.Image can be read with .open or created directly from an numpy array
(maybe read with np.loadtxt)
import jscatter as js import PIL import numpy as np image0 = PIL.Image.open(js.examples.datapath+'/calibration.tiff') # read image imagearray = np.random.rand(256,256)*100 # array image1 = PIL.Image.fromarray(imagearray) # PIL image (try image1.show()) # create sasImage with needed parameters # from a PIL image si=js.sas.sasImage(image1,1,[100,100],pixel_size=0.001,wavelength=2) # or directly from numpy arra sa=js.sas.sasImage(imagearray,1,[100,100],pixel_size=0.001,wavelength=2) sa.show()
References
[1]Everything SAXS: small-angle scattering pattern collection and correction Brian Richard Pauw J. Phys.: Condens. Matter 25, 383201 (2013) DOI https://doi.org/10.1088/0953-8984/25/38/383201
[2]FabIO: easy access to two-dimensional X-ray detector images in Python E. B. Knudsen, H. O. Sørensen, J. P. Wright, G. Goret and J. Kieffer Journal of Applied Crystallography, Volume 46, Part 2, pages 537-539
Examples
Read TIFF images from our Ganesha Instrument which includes all needed data for processing in the tiff tags.
The procedure of calibration, masking areas, radial averaging and more is shown.
import jscatter as js # # Look at calibration measurement calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') # Check center # For correct center it should show straight lines (change center to see change) calibration.showPolar(center=calibration.center,scaleR=3) # or use pickBeamcenter which seems to be more accurate calibration.pickBeamcenter() # Recalibrate with previous found center (calibration sets it already) calibration.recalibrateDetDistance(showfits=True) iqcal=calibration.radialAverage() # This might be used to calibrate detector distance for following measurements as # empty.setDetectorDistance(calibration) # empty = js.sas.sasImage(js.examples.datapath+'/emptycell.tiff') # Mask beamstop (not the same as calibration, unluckily) empty.mask4Polygon([370,194],[380,194],[466,0],[456,0]) empty.maskCircle(empty.center, 17) empty.show() buffer = js.sas.sasImage(js.examples.datapath+'/buffer.tiff') buffer.maskFromImage(empty) buffer.show() bsa = js.sas.sasImage(js.examples.datapath+'/BSA11mg.tiff') bsa.maskFromImage(empty) bsa.show() # by default a log scaled image # # subtract buffer (transmission factor is just a guess here, sorry) new=bsa-buffer*0.2 new.show() # iqempty=empty.radialAverage() iqbuffer=buffer.radialAverage() iqbsa=bsa.radialAverage() # p=js.grace(1,1) p.plot(iqempty,le='empty cell') p.plot(iqbuffer,le='buffer') p.plot(iqbsa,le='bsa 11 mg/ml') p.title('raw data, no transmission correction') p.yaxis(min=1,max=1e3,scale='l',label='I(q) / a.u.') p.xaxis(scale='l',label='q / nm\S-1') p.legend()
Reading various X-ray detector formats In the second example reading of an exemplary EDF file from ALBA is shown. Here the needed information is not stored in the file.
Dependent on the used instrument many different names for the needed parameters are used.
Contact your instrument responsible to get the respective information or which name is used in the file. The below used file contains the energy_bragg parameter with the corresponding Xray energy.
import jscatter as js import glob # read edf calibration file (internally fabio is used for reading) calibration = js.sas.sasImage(js.examples.datapath+'/calibration.edf.gz') # add parameters defining detector data (here wavelength) according to detector data in header energy = calibration.energy_bragg[0] lam = 6.625E-34 * 3E8 / (1.6E-19* energy * 1000) * 1E10 calibration.setWavelength(lam) calibration.setPixelSize([0.000172,0.000172]) calibration.setDetectorDistance(2.5) # calibration.pickBeamcenter() # needed to detect center calibration.setPlaneCenter([491.08,570.66]) # if known from above line calibration.recalibrateDetDistance(showfits=1) # now treat the data files=glob.glob('*.edf') for file in files: # option *copy* copies parameters from calibration above image = js.sas.sasImage(file,copy=calibration) # treat data as you need........
- property array¶
Strip of all attributes and return a simple array without mask.
- asImage(scale='log', colormap='jet', inverse=False, linthresh=1.0, linscale=1.0)[source]¶
Returns the sasImage as 8bit RGB image using PIL.
See PIL(Pillow) for more info about PIL images and image manipulation possibilities as e.g. in notes. Conversion to 8bit RGB looses floating point information but is for presenting and publication.
- Parameters:
- scale‘log’, ‘symlog’, default = ‘norm’
Scale for intensities.
‘norm’ Linear scale.
‘log’ Logarithmic scale
‘symlog’ Symmetrical logarithmic scale is logarithmic in both the positive and negative directions from the origin. Use linthresh, linscale to adjust.
- colormapstring, None
Colormap from matplotlib or None for grayscale. For standard colormap names look in js.mpl.showColors().
- inversebool
Inverse colormap.
- linthreshfloat, default = 1
Only used for scale ‘sym’. The range within which the plot is linear (-linthresh to linthresh).
- linscalefloat, default = 1
Only used for scale ‘sym’. Its value is the number of decades to use for each half of the linear range.
- Returns:
- PIL image
Notes
Pillow (fork of PIL) allows image manipulation. As a prerequisite of Jscatter it is installed on your system and can be imported as
import PILimage=mysasimage.asImage() image.show() # show in system default viewer image.save('test.pdf', format=None, **params) # save the image in different formats image.save('test.jpg',subsampling=0, quality=100) # use these for best jpg quality image.save('test.png',transparency=(0,0,0)) # png image with black as transparent image.crop((10,10,200,200)) # cut border import PIL.ImageOps as PIO nimage=PIO.equalize(image, mask=None) # Equalize the image histogram. nimage=PIO.autocontrast(image, cutoff=0, ignore=None) # Automatic contrast nimage=PIO.expand(image, border=20, fill=(255,255,255)) # add border to image (here white)
Examples
import jscatter as js import PIL.ImageOps as PIO cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') # create image for later usage image=cal.asImage(colormap='inferno',scale='log',inverse=1) # create image and just show it cal.asImage(colormap='inferno',scale='log').show() # expand image and show it or save it PIO.expand(image, border=20, fill=(255,255,255)).show() PIO.expand(image, border=20, fill=(255,255,255)).save('myimageas.pdf')
- asdataArray(masked=0)[source]¶
Return representation of sasImage as dataArray with (qx, qy, qz, I(qx,qy,qz)).
- Parameters:
- maskedfloat, None, string, default=0
- How to deal with masked values.
float : Set masked pixels to this value
- NoneRemove from dataArray.
To recover the image the masked pixels need to be interpolated on a regular grid.
- ‘linear’, ‘cubic’, ‘nearest’interpolate masked points by scipy.interpolate.griddata
using specified order of interpolation on 2D image.
‘radial’ Use the radial averaged data to interpolate.
- Returns:
- dataArray with [qx, qy, qz, I(qx,qy,qz)]
.qx, .qy : original q pixel values corresponding to image pixels along axes through the center to recover the image. (.qx for .qy=0 and .qy for .qx=0.) Please keep in mind that the values are only exact for integer center values. Values are also not equidistant as for larger values the curvature of the Ewald sphere is important. To recover the image use .regrid(method=’nearest’) to avoid artefacts due to this inaccuracy and mask values according to original mask.
.sasImageshape as shape of original sasImage.
Image attributes except [‘artist’, ‘imageDescription’] are copied.
[qx,qy,qz] correspond to [.X, .Z, .W] in dataArray with .Y as I(qx,qy,qz).
The third dimension .qz (.W in dataArray) results from the fact that the flat detector image represents the scattering vectors on the Ewald sphere which has also a qz component.
For small angle scattering this component might be small compared to qx,qy.
Examples
Use radial averaged data to interpolate the regions where the detector is dark
import jscatter as js import numpy as np bsa = js.sas.sasImage(js.examples.datapath+'/BSA11mg.tiff') bsa.maskCircle(bsa.center,40) bsar=bsa.asdataArray('radial') js.mpl.surface(bsar.X, bsar.Z, bsar.Y,shape=bsar.sasImageshape)
This demo will show the interpolation in the masked regions of an artificial intensity distribution. The examples might allow interpolation in a masked region like a unwanted Bragg reflex:
import jscatter as js import numpy as np calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') # manipulate data (not the mask) # this only creates here a flat plateau for later interpolation. calibration.data[:300,60:1200]=100 calibration.data[:300,120:180]=300 calibration.data[:300,180:]=600 # mask a circle calibration.maskCircle([200,200], 120) cal=calibration.asdataArray('linear') cal.Y[cal.Y<=0.1]=1.1 js.mpl.surface(cal.X, cal.Z, cal.Y,shape=cal.sasImageshape) cal2=calibration.asdataArray(None) # this is reduced in size due to the mask
- azimuthAverage(center=None, qrange=[None, None], number=180, kind='lin', calcError=False)[source]¶
Azimuthal average of image and conversion to wavevector q.
Remember to set .detector_distance to calibrated value.
- Parameters:
- centerlist 2x float
Sets center in data and uses this. If not given the attribute center in the data is used.
- qrange2x float, default [0,max]
Range of q values to include as [min,max].
- numberint, default 180
Number of intervals on new X scale.
- kind‘log’, default ‘lin’
Determines how points are distributed.
- calcError‘poisson’,’std’, default None
- How to calculate error.
- ‘poisson’ according to Poisson statistics.
Use only for original images showing unprocessed photon counts.
‘std’ as standard deviation of the values in an interval.
otherwise no error
- Returns:
- dataArray with added attributes of the image. artist and entriesXML are ommited
Notes
Correction of pixel size for flat detector projected to Ewald sphere included.
The value in a q binning is the average count rate \(c(q)=(\sum c_i)/N\) with counts in pixel i \(c_i\) and number of pixels \(N\)
calcError : If the image is unprocessed (no background subtraction or transmission correction) containing original photon count rates the standard error can be calculated from Poisson statistic.
The error (standard deviation) is calculated in a q binning as \(e=(\sum c_i)^{1/2}/N\)
The error is valid for single photon counting detectors showing Poisson statistics as the today typical Pilatus detectors from DECTRIS.
The error for \(\sum c_i) <= 0\) is set to zero. One may estimate the corresponding error from neighboring intervals.
In later 1D processing as e.g. background correction the error can be included according to error propagation.
‘std’ calcs the error as standard deviation in an interval.
Examples
import jscatter as js cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') p=js.grace() az=cal.azimuthAverage(qrange=[0.9,1.3]) p.plot(az,legend='q=0.9-1.3 nm\S-1') az=cal.azimuthAverage(qrange=[2,2.4]) p.plot(az,legend='q=2-2.4 nm\S-1') az=cal.azimuthAverage(qrange=[3,3.5]) p.plot(az,legend='q=3-3.5 nm\S-1') az=cal.azimuthAverage(qrange=[4.1,4.4]) p.plot(az,legend='q=4.1-4.4 nm\S-1') p.yaxis(label=r'I(\xj\f{})',scale='log') p.xaxis(label=r'azimuth \xj\f{} / rad') p.title('The AgBe is not isotropically ordered') p.legend(x=0,y=1) p.text('beamstop arm',x=-2,y=0.1) p.text('mask',x=-3,y=4)
- calibrateOffsetDetector(lattice=None, center=None, distance=None, alpha=None, beta=None, gamma=None, domainsize=1000, asym=0, lg=1, rmsd=0.02, hklmax=17)[source]¶
Compare sasImage to calibration standard in powder average to determine detector position.
Any detector orientation relative to the sample and incoming beam can be used to calibrate parameters describing the detector position. A standard sample as silicon (large angles) or AgBe (small angles) can be used. Opens a window with changeable parameters to align sasImage and simulated lattice image. This can also be used in standard SAS geometry with the detector normal being the incoming beam direction.
- Parameters:
- latticelattice object
A lattice object (see structurefactor lattices) representing the used reference material to determine the expected scattering pattern. E.g.
silicon = js.sf.diamondLattice(0.543, 1)- center,distance,alpha,beta,gamma2x float, float, float,float
Parameters determining the detector position. See
sasImage.setDetectorPosition()for detailed description.- domainsizefloat
Domainsize of the used powder changing the peak width. See
latticeStructureFactor().- asymfloat
Asymmetry of the peaks. See
latticeStructureFactor().- lgfloat
Lorenzian/gaussian fraction. See
latticeStructureFactor().- rmsdfloat
Root mean square displacement in lattice . See
latticeStructureFactor().- hklmaxint, default = 17
Maximum order of the Bragg peaks to include. If hklmax is to small bragg peaks of higher order might be missing.
- Returns:
- None
- Sets corresponding values in setDetectorPosition(…) when window is closed.
Notes
Usage: - Opens a window with
Original gray scale image of calibration measurements with colored overlay for simulated peak positions.
A radial average over the image and simulated data. The radial average depends on center location and angles and is updated if parameters change.
The overlay in left image shows the range between selected doted lines in right image. - The selection is done using the right/left mouse button for max/min values. - Selection might highlight the maxima of peaks or the flanks dependent on the selected range.
Small ranges highlight thin lines in the 2D image.
Lines have to be aligned to measured pattern and peak positions need to coincident.
Use the calibrated sasImage as argument to copy while creating a new sasImage to use (copy) calibrated values in new image.
A detector geometry as 45deg with the beamcenter at the detector edge might be used to cover a large region from lowest to highest wavevectors.
Examples
Silicon powder for an offset detector located parallel to the incoming beam in reverse orientation (beta=90) The detector was positioned about 70 mm away from the incoming beam a bit behind the sample. Here the distance and center need to be adjusted.
import jscatter as js sic = js.sas.sasImage(js.examples.datapath+'/Silicon.tiff') silicon = js.sf.diamondLattice(0.543, 1) cc=[130,-100] sic.setPlaneOrientation(90,90,0) sic.calibrateOffsetDetector(center=cc,distance=0.070,lattice=silicon,domainsize=30,rmsd=0.003)
Conventional AgBe reference with center located on the detector. Beamcenter is center on detector.
import jscatter as js # cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') # mask some parts of the detector (because of shading and the beam stop) to get clearer radial average. bc=cal.center # beamstop arm cal.mask4Polygon([bc[0]+5,bc[1]],[bc[0]-5,bc[1]],[bc[0]-5+43,0],[bc[0]+5+43,0]) # beamstop cal.maskCircle(cal.center, 18) # shade of beam entrance cal.maskCircle([500,320], 280,invert=True) # AgBe reference agbe=js.sf.latticeFromCIF(js.examples.datapath+'/1507774.cif',size=[1,1,1]) cal.calibrateOffsetDetector(center=cal.center,distance=0.18,lattice=agbe,domainsize=50,rmsd=0.003)
- findCenterOfIntensity(center=None, size=100)[source]¶
Find beam center as center of intensity..
Only values above the mean value are used to calc center of intensity. Use an image with a clear symmetric and strong scattering sample as AgBe or empty beam measurement. Use .showPolar([600,699],scaleR=5) to see if peak is symmetric.
- Parameters:
- centerlist 2x int
First estimate of center as [height, width] position. If not given preliminary center is estimated as center of intensity of full image.
- sizeint
Defines size of rectangular region of interest (ROI) around the center to look at.
- Returns:
- Adds (replaces) .center as attribute.
Notes
If ROI is to large the result may be biased due to asymmetry of the intensity distribution inside of ROI.
Additional strong masking in ROI leads to bias of the found center.
- gaussianFilter(sigma=2)[source]¶
Gaussian filter in place.
Uses ndimage.filters.gaussian_filter with default parameters except sigma.
- Parameters:
- sigmafloat
Gaussian kernel sigma.
- getPolar(center=None, scaleR=1, offset=0)[source]¶
Transform to polar coordinates around center with interpolation.
Azimuth corresponds: center line upwards, upper quarter center to right upper/lower edge = center downwards, lower quarter center to left
- Parameters:
- center[int,float]
Beamcenter
- scaleRfloat
Scaling factor for radial component to zoom. Works only for alpha,beta,gamma = 0 .
- offsetfloat >0
Offset to remove center from polar image. Works only for alpha,beta,gamma = 0 .
- Returns:
- ndarray
Examples
See showPolar for examples.
Use radial averaged data to interpolate
import jscatter as js calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') pol = calibration.getPolar()
- getfromcomment(name, replace=None, newname=None)[source]¶
Extract name from .artist or .imageDescription with attribute name in front.
If multiple names start with parname first one is used. Used line is deleted from .artist or .imageDescription.
- Parameters:
- namestring
Name of the parameter in first place.
- replacedict
Dictionary with pairs to replace in all lines.
- newnamestring
New attribute name
- property iX¶
X pixel coordinates
- property iY¶
Y pixel coordinates
- interpolateMaskedRadial(radial=None)[source]¶
Interpolate masked values from radial averaged image or function.
This can be used to “extrapolate” over masked regions if e.g a background was measured at wrong distance.
- Parameters:
- radialdataArray, function, default = None
- Determines how to determine masked values based on radial q values from center.
Function accepting array to calculate masked data.
dataArray for linear interpolating masked points.
None uses the radialAverage image.
- Returns:
- sasImage including original parameters.
Examples
Use radial averaged data to interpolate
import jscatter as js import numpy as np calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') cal=calibration.interpolateMaskedRadial() # or # cal=calibration.interpolateMaskedRadial(calibration.radialAverage()) cal.show()
Generate image for different detector distance
cal.setDetectorDistance(0.3) # mask whole image cal.mask=np.ma.masked # recover image with radial average from original cal2=cal.interpolateMaskedRadial(calibration.radialAverage()) cal2.show()
- lineAverage(sigma=3, darkline=None, whiteline=None)[source]¶
Line average and conversion to wavevector q for Kratky (line) collimation cameras.
Dark current subtraction, cosmic spikes and dead pixel correction are applied before line average. Remember to set .detector_distance to calibrated value.
- Parameters:
- darklinedataArray
lineAverageof a dark current measurement. White pixel are present in dark measurement and corrected by subtracting the dark current. This is subtracted before any later processing.- sigmafloat, default 3
sigma is factor for standard deviation. Singular peaks (from spikes in time due to cosmic rays) are filtered out from the time series if spike counts are
abs(count - mean) > sigma * std. If 0 no filter is applied.- whitelinedataArray
lineAverage of whiteline as a const scattering like empty cell or buffer (after dark correction).
Dead pixel with zero count lead to a drop in counts for a single pixel. An approximate scaling factor is evaluated from the difference to the neighbor average to account for the missing dead pixels.
- Returns:
- dataArray
.filename
.detector_distance
.description
.center
Notes
detector_distance in attributes is used for scattering vector calculation.
.center of the primary beam needs to be defined using
lineFindCenter().Correction for flat detector projected to Ewald sphere included.
For Kratky camera setups as SAXSpace from Anton Paar using a CCD camera. Each column in the saved image corresponds to a timeframe (e.g. 10s counting) that is averaged over a horizontal pixel line on the CCD chip.
White pixel lead to values above vertical neighbor average. These are detected from a dark measurement as they are present also there. Pixel proper subtraction corrects dark readout noise and white pixels.
Dead pixel lead to values lower than neighbor average. These are detected from a nearly flat measurement as empty cell or water measurement. Values are corrected according to a pixel scaling factor close to N/n for n dead pixels in a row of N pixels.
Examples
Take line average over Kratky camera image
Additional split image as time series. From the first full average the center is determined with higher accuracy and used in later series. E.g. to check sample stability (aggregation) or sedimentation in the sample.
import jscatter as js import numpy as np # for later desmearing beam = js.sas.readpdh(saxspace+'BeamProfile.pdh') mbeam = js.sas.prepareBeamProfile(beam, bxw=0.013, dIW=1.33) # empty cell as whiteline iempty = js.sas.sasImage(saxspace+'emptycell2_T10_360Frames.tif',flip=1) iempty.lineFindCenter([20,80]) iempty.setDetectorDistance(0.303) # dark measurement for dark count correction idark = js.sas.sasImage(saxspace+'Dark Current_360Frames.tif',flip=1) idark.center = iempty.center dark = idark.lineAverage(sigma=3) # use empty cell to generate whiteline for dead pixel correction with dark correction whiteline = iempty.lineAverage(darkline=dark) # recalculate empty cell with dark count and whiteline correction empty = iempty.lineAverage(darkline=dark,whiteline=whiteline) # treat all measurements in the same way ibuff = js.sas.sasImage(saxspace+'buffer2_T10_305mm_360Frames.tif',flip=1) ibuff.setDetectorDistance(0.303) ibuff.center = iempty.center buff = ibuff.lineAverage(darkline=dark,whiteline=whiteline)
Comparison of time evolution e.g. for radiation damage or sedimentation.
p=js.grace() p.plot(buff) step = 10 for t in np.r_[:image.shape[0]:step]: data = ibuff[t:t+step].lineAverage(darkline=dark,whiteline=whiteline) p.plot(data)
- lineFindCenter(minmax=[2, 80], use='COM', show=False, darkline=None)[source]¶
Find center of primary beam for line collimation cameras with semitransparent beamstop.
- Parameters:
- minmax[int,int], default [2,50]
Interval for determination of center in pixel units.
- use‘com’,’gauss’, default ‘com’
Select center of mass (‘com’) of the peak or center of Gaussian fit.
- showbool
Show the Gaussian fit of the primary beam.
- darklinedataArray
lineAverage of dark current measurement. This is subtracted before any processing and should have same counting-time as sasImage.
Notes
Adds center, primarybeam_hwhm, primarybeam_peakmax attributes.
The primary beam is detected using a Gaussian fit to the peak between min-max.
- mask4Polygon(p1, p2, p3, p4, invert=False)[source]¶
Mask inside polygon of 4 points.
Points need to be given in right hand order.
- Parameters:
- p1,p2,p3,p4list of 2x float
Edge points.
- invertbool
Invert region. Mask outside circle.
- maskCircle(center, radius, invert=False)[source]¶
Mask points inside circle.
- Parameters:
- centerlist of 2x float
Center point. This is not the plane center.
- radiusfloat
Radius in pixel units
- invertbool
Invert region. Mask outside circle.
- maskFromImage(image)[source]¶
Use/copy mask from image.
- Parameters:
- imagesasImage
sasImage to use mask for resetting mask. image needs to have same dimension.
- maskRegion(xmin, xmax, ymin, ymax)[source]¶
Mask rectangular region.
- Parameters:
- xmin,xmax,ymin,ymaxint
Corners of the region to mask
- maskRegions(regions)[source]¶
Mask several regions.
- Parameters:
- regionslist
List of regions as in maskRegion.
- maskReset()[source]¶
Reset the mask.
By default values smaller 0 are automatically masked again as is also default for reading
- maskSectors(angles, width, radialmax=None, invert=False)[source]¶
Mask sector around center.
Zero angle is
- Parameters:
- angleslist of float
Center angles of sectors in grad.
- widthfloat or list of float
Width of the sectors in grad. If single value all sectors are equal.
- radialmaxfloat
Maximum radius in pixels.
- invertbool
Invert mask or not.
Examples
import jscatter as js cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') cal.maskSectors([-90,0,90,-180],20,radialmax=200,invert=True) cal.show()
- maskTriangle(p1, p2, p3, invert=False)[source]¶
Mask inside triangle.
- Parameters:
- p1,p2,p3list of 2x float
Edge points of triangle.
- invertbool
Invert region. Mask outside circle.
- maskbelowLine(p1, p2)[source]¶
Mask points at one side of line.
The masked side is left looking from p1 to p2.
- Parameters:
- p1, p2list of 2x float
Points in pixel coordinates defining line.
- property pQ¶
3D scattering vector \(q\) for pixels with detector placed in standard SAS or offset geometry.
The detector is placed at an offset position with the detector normal rotated against the incoming beam direction by angles alpha, beta, gamma. If these are zero we have conventional SAS geometry.
Use .setDetectorPosition to place the detector.
- Returns:
- array
- scattering vector with shape(image) x 3 in cartesian coordinates and units 1/nm.
Notes
The incident beam is directed in Z direction \(k_{i}=[0, 0, k]\).
The scattered wavevector is
\[\begin{split}k_f=k \begin{bmatrix} cos(\phi)sin(\theta) \\ sin(\phi)sin(\theta) \\ cos(\theta) \end{bmatrix} = \begin{bmatrix} k_x \\ k_y \\ k_z \end{bmatrix}\end{split}\]with polar coordinates \(k, \phi, \theta\) where \(\theta\) is the conventional scattering vector used in small angle scattering.
The scattering vector is
\[\begin{split}q=k_f-k_i= k \begin{bmatrix} cos(\phi)sin(\theta) \\ sin(\phi)sin(\theta) \\ cos(\theta) -1 \end{bmatrix}\end{split}\]with \(|q|=4\pi/\lambda\) and \(|k|=2\pi/\lambda\).
Pixel and pQ orientation
- The convention is as follows
Pixel origin [0,0] is upper left corner with coordinates in [height, width] of the detector.
pQ (wavevector coordinates) are that the pixel origin [0,0] is in the [positive, negative] quadrant that the lower left corner is [negative,negative] as expected for a scattering pattern with axes from left to right and bottom up and the origin (incident beam) somewhere in the image and viewed from the sample location.
- pQaxes()[source]¶
Get scattering vector along detector pixel axes X, Y around center.
In standard small angle geometry the detector pixel X,Y directions are perpendicular to the incident beam in Z direction. For an offset detector this not necessarily the case as we have curvilinear coordinates. Needs wavelength, detector_distance and center defined.
- Returns:
- qx,qy with image x and y dimension
- property pQnorm¶
3D scattering vector \(|q|\) for detector pixel. See .pQ .
- pickBeamcenter(levels=8, symmetry=6)[source]¶
Pick the beam center from a calibration sample as AgBe in standard SAS geometry if a beamstop is used.
For measurements without beamstop use
sasImage.findCenterOfIntensity()with an arbitrary image.If the beamstop covers the beam pickBeamcenter uses radial averaged sectors to find the optimal center with best overlap of peaks in the sectors. Closing the image accepts the actual selected center. Standard SAS geometry => alpha=beta=gamma=0
- Parameters:
- levelsint
Number of levels in contour image.
- symmetryint
Number of sectors around center for radial averages.
- Returns:
- After closing the selected center is saved in the sasImage.
Notes
How it works A figure with the AgBe picture (right) and a radial average over sectors is shown (left, symmetry defines number of sectors) .
Beamcenter: A circle is shown around the center. Mouse left click changes the center to mouse pointer position.
The center can be moved by arrow keys (+-1) or ctrl+arrow (+-0.1)
The default radius corresponds to an AgBe reflex. By middle or right click the radius can be set to mouse pointer position. Additional the radius of the circle (center of left plot data) can be increased/decreased by +/-.
Width around radius (for left plot) can be increased/decrease by ctrl++/ctrl+-.
A radial average in sectors is calculated (after some smoothing) and shown in the left axes.
The center is OK if the peaks show maximum overlap and symmetry.
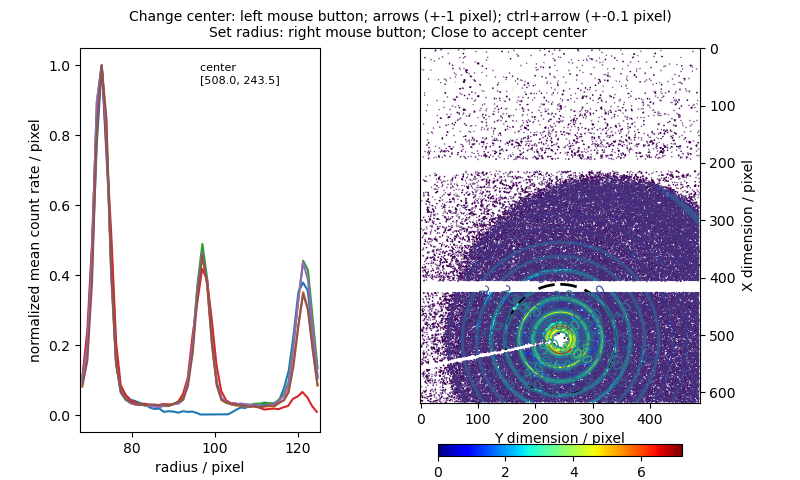
Examples
import jscatter as js # calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') # use pickBeamcenter calibration.pickBeamcenter()
- radialAverage(center=None, number=300, kind='log', calcError=False, units=None)[source]¶
Radial average of image and conversion to wavevector q.
Remember to set .detector_distance to calibrated value. Setting units to ‘sr’ will scale the output to counts/micro_steradians which also accounts for different detector distances.
- Parameters:
- centerlist 2x float
Sets center in data and uses this. If not given the attributecenter in the data is used.
- numberint, default 500
Number of intervals on new X scale.
- kind‘lin’, ‘log’, array default ‘log’
Determines how points are distributed on the Q scale. (See dataArray.prune) It may be an array with an explicit list from another radialAveraged image (like im.radialAverage(…).X) . If the images have different centers then some intervals around the center might be empty. These are filled with interpolated values. Just avoid different centers.
- calcError‘poisson’,’std’, default None
- How to calculate error.
- ‘poisson’ according to Poisson statistics.
Use for original images showing unprocessed photon/neutron counts. E.g for Dectris detectors or SANS detectors, but not for CCD cameras.
‘std’ as standard deviation of the values in an interval.
otherwise no error
- unitsNone, ‘sr’
- Units of the returned radial average as :
None default counts per pixel
‘sr’ counts per solid angle as counts/micro steradians = 1e-6 counts/steradians. This accounts also different detector distances.
- Returns:
- dataArray with added attributes of the image. artist and entriesXML are ommited
Notes
Correction of pixel size for flat detector projected to Ewald sphere included. The correction is relative to the pixel located at the center. The intensity remains in units counts/pixel.
The value in a q binning is the average count rate \(c(q)=(\sum c_i)/N\) with counts in pixel i \(c_i\) and number of pixels \(N\)
Setting units to ‘sr’ scales to counts per solid angle (micro steradians). In these units the scattered intensity is independent of the detector distance. Scaling is done after calculation of the error.
calcError : If the image is unprocessed (no background subtraction or transmission correction) containing original photon count rates the standard error can be calculated from Poisson statistic.
The error (standard deviation) is calculated in a q binning as \(e=(\sum c_i)^{1/2}/N\)
The error is valid for single photon counting detectors showing Poisson statistics as the today typical Pilatus detectors from DECTRIS.
The error for \(\sum c_i) <= 0\) is set to zero. One may estimate the corresponding error from neighboring intervals.
In later 1D processing as e.g. background correction the error can be included according to error propagation.
‘std’ calcs the error as standard deviation in an interval.
Examples
For Pilatus detector including some average with less pixel at high Q I use
sample = image.radialAverage(kind='log',number=200, calcError='poisson',units='sr').prune(0.1,6)
Mask and do radial average over sectors.
import jscatter as js cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') p=js.grace() calc=cal.copy() calc.maskSectors([0,180],20,radialmax=100,invert=True) calc.show() icalc=calc.radialAverage() p.plot(icalc,le='horizontal') calc=cal.copy() calc.maskSectors([90+0,90+180],20,radialmax=100,invert=True) calc.show() icalc=calc.radialAverage() p.plot(icalc,le='vertical') p.yaxis(scale='l') p.legend() p.title('The AgBe is not isotropically ordered')
- recalibrateDetDistance(center=None, number=500, fcenter=1.0, fwhm=0.1, peaks=None, showfits=False)[source]¶
Recalibration of detectorDistance by AgBe reference for point collimation.
Use only for AgBe reference measurements to determine the correction factor. For non AgBe measurements set during reading or .detector_distance to the new value. May not work if the detector distance is totally wrong.
- Parameters:
- centerlist 2x float
Sets beam center or radial center in data and uses this. If not given the attributecenter in the data is used.
- numberint, default 1000
number of intervals on new X scale.
- fcenterfloat, default 1
Determines start value for peak fitting.
By default, the position of the peak maximum is used if it is larger than (mean(Y)+2std(Y)) of the signal Y. Otherwise fcenter*peakposition[i] is used. Negative fcenter forces the start value to be |fcenter|*peakposition[i].
- Reference peakpositions in 1/nm:
[1.0753, 2.1521, 3.2286, 4.3049, 5.3813, 6.4576, 7.5339, 8.6102, 9.6865, 10.7628]
- fwhmfloat, default 0.1
Start value for full width half maximum in peak fitting.
- peakslist of float
Peak positions of a reference measurement. By default these are the AgBe peaks. Check
`js.sas.AgBepeaks`. Can also be used to limit the used peaks if one is bad or change the reference material.- showfitsbool
Show the AgBe peak fits.
Notes
.distanceCorrection will contain factor for correction. Repeating this results in a .distanceCorrection close to 1.
To check the result use showfits=True.
The fits should all fit well to accept the result. To improve use
sasImage.maskCircle()to cover an inside region and withinvert=Truethe outside edges. For bad result try to change the detectordistance to a more reasonable value as first guess. This can be recognized if the AgBe peaks shown with showfits=True are not approximately centered within the data range for fitting.
We fit a Voigt function to each of the detected peaks in the image and use the average of the resulting correction factors for each peak as overall correction factor.
As a background we fit a power law as this is needed on some beam line machines.
- reduceSize(bin=2, center=None, border=None)[source]¶
Reduce size of image using uniform average in box.
Center, pixel_size are scaled correspondingly.
- Parameters:
- binint
Size of box to average within. Also factor for reduction in image size.
- center[int,int]
Center of crop region.
- borderint
- Size of crop region.
If center is given a box with 2*size around center is used.
If center is None the border is cut by size.
- Returns:
- sasImage
- saveAsTIF(filename, fill=None, **params)[source]¶
Save the sasImage as float32 tif without loosing information.
Conversion from float64 to float32 is necessary. To save colored images use asImage.save() (see
asImage())- Parameters:
- filenamestring
Filename to save to.
- fillfloat, ‘min’ default None
Fill value for masked values. By default this is -1.
- ‘min’ uses the minimal value of the respective data type
np.iinfo(np.int32).min = -2147483648 for int32
np.finfo(np.float32).min = -3.4028235e+38 for float32
- paramskwargs
Additional kwargs for PIL.Image.save if needed.
Examples
import jscatter as js cal = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') cal2=cal/2. cal2.saveAsTIF('mycal',fill=-100) mycal=js.sas.sasImage('mycal.tif',maskbelow=-200) mycal.show()
- setAttrFromImage(image)[source]¶
Copy center, detector_distance, alpha, beta, gamma wavelength, pixel_size from image.
- Parameters:
- image sasImage
sasImage to copy attributes to self.
- setDetectorDistance(detector_distance)[source]¶
Set detector distance as shortest distance to sample along plane normal vector.
- Parameters:
- detector_distancefloat, sasImage
New value for detector distance in units m. If sasImage the detector_distance is copied.
Notes
EXIF data show this as list so we stay to this.
- setDetectorPosition(center, detector_distance, alpha=None, beta=None, gamma=None)[source]¶
Set parameters describing the position and orientation of the detector.
- Parameters:
- center2x float
Center of the detector where the plane normal is going through the sample origin in pixel units. For conventional small angle scattering geometry (detector plane perpendicular to incoming beam) this is the beam center.
- detector_distancefloat
Distance of the detector center to sample origin in units m.
- alphafloat
Angle between incident beam and detector plane normal in degree. Or the angle between incident beam and the vector pointing from the sample to the detector center. E.g. 0° for SAS.
- betafloat
Rotation angle of detector pixel X dimension around detector plane normal in degree. Determines orientation of detector, rotating the detector in plane.
- gammafloat
Rotation of detector normal around incident beam in degree. Position of the detector as seen from sample or like rotating the sample around incident beam.
For Debye-Scherer ring pattern (powder average) equal 0.
- setPixelSize(pixel_size)[source]¶
Set pixel_size.
- Parameters:
- pixel_size[float,float]
Pixel size in [x,y] direction in units m.
- setPlaneCenter(center)[source]¶
Set beamcenter or center of detector plane where plane normal has shortest distance to sample.
In standard SAS geometry this is equal to the beamcenter. For offset detectors is point of closest distance.
- Parameters:
- center2x float, sasImage
New value for center as [height, width] coordinates. If sasImage the center is copied.
- setPlaneOrientation(alpha=None, beta=None, gamma=None)[source]¶
Set orientation angles of detector plane .
In standard SAS geometry these are equal 0.
- Parameters:
- alphafloat
Angle between incident beam and detector plane normal in degree. Or the angle between incident beam and the vector pointing from the sample to the detector center. E.g. 0° for SAS.
- betafloat
Rotation angle of detector pixel X dimension around detector plane normal in degree. Determines orientation of detector, rotating the detector in plane.
- gammafloat
Rotation of detector normal around incident beam in degree. Position of the detector as seen from sample or like rotating the sample around incident beam.
For Debye-Scherer ring pattern (powder average) equal 0.
- setWavelength(wavelength)[source]¶
Set wavelength.
- Parameters:
- wavelength[float]
Wavelength in units angstrom.
- show(**kwargs)[source]¶
Show sasImage as matplotlib figure.
- Parameters:
- scale‘log’, ‘symlog’, default = ‘norm’
Scale for intensities.
‘norm’ Linear scale.
‘log’ Logarithmic scale
‘symlog’ Symmetrical logarithmic scale is logarithmic in both the positive and negative directions from the origin. This works also for only positive data. Use linthresh, linscale to adjust.
- levelsint, None
Number of contour levels.
- colorMapstring
Get a colormap instance from name. Standard mpl colormap name (see showColors).
- badcolorfloat, color
Set the color for bad values (like masked) values in an image. Default is bad values be transparent. Color can be matplotlib color as ‘k’,’b’ or float value in interval [0,1] of the chosen colorMap. 0 sets to minimum value, 1 to maximum value.
- linthreshfloat, default = 1
Only used for scale ‘symlog’. The range within which the plot is linear (-linthresh to linthresh).
- linscalefloat, default = 1
Only used for scale ‘symlog’. Its value is the number of decades to use for each half of the linear range. E.g. 10 uses 1 decade.
- lineMapstring
Label color Colormap name as in colorMap, otherwise as cs in in Axes.clabel * if None, the color of each label matches the color of the corresponding contour * if one string color, e.g., colors = ‘r’ or colors = ‘red’, all labels will be plotted in this color * if a tuple of matplotlib color args (string, float, rgb, etc),
different labels will be plotted in different colors in the order specified
- fontsizeint, default 10
Size of line labels in pixel
- axisNone, ‘pixel’
If coordinates should be forced to pixel, otherwise wavevectors if possible.
- invert_yaxis, invert_xaxisbool
Invert corresponding axis.
- blockbool
Open in blocking or non-blocking mode
- origin‘lower’,’upper’
Origin of the plot. See matplotlib imshow.
- Returns:
- image handle
Notes
To show data as Image in correct orientation the option ax.invert_yaxis() is used. If you plot directly with matplotlib use the same.
Examples
Use radial averaged data to interpolate
import jscatter as js import numpy as np calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') calibration.show(colorMap='ocean') calibration.show(scale='sym',linthresh=20, linscale=5)
Use
scale='symlog'for mixed lin-log scaling to pronounce low scattering. See mpl.contourImage for more options also available using.show.import jscatter as js import numpy as np # sets negative values to zero bsa = js.sas.sasImage(js.examples.datapath+'/BSA11mg.tiff') fig=js.mpl.contourImage(bsa,scale='sym',linthresh=30, linscale=10) fig.axes[0].set_xlabel(r'$Q_{{ \mathrm{{X}} }}\;/\;\mathrm{{nm^{{-1}}}}$ ') fig.axes[0].set_ylabel(r'$Q_{{ \mathrm{{Y}} }}\;/\;\mathrm{{nm^{{-1}}}}$ ')
- showPolar(center=None, scaleR=1, offset=0, scale='log')[source]¶
Show image transformed to polar coordinates around center.
Azimuth for standard SAS geometry corresponds to: center line upwards, upper quarter center to right upper/lower edge = center downwards, lower quarter center to left
- Parameters:
- center[int,int]
Beamcenter
- scaleRfloat
Scaling factor for radial component to zoom the center. Works only for alpha,beta,gamma = 0 .
- offsetfloat
Offset to remove center from polar image. Works only for alpha,beta,gamma = 0 .
- scale‘log’, ‘symlog’, default = ‘log’
Scale for intensities.
‘norm’ Linear scale.
‘log’ Logarithmic scale
‘symlog’ Symmetrical logarithmic scale is logarithmic in both the positive and negative directions from the origin. This works also for only positive data. Use linthresh, linscale to adjust.
- Returns:
- Handle to figure
Examples
Use polar coordinates to see if center is in middle of Debye-Scherrer rings. First standard SAS geometry:
import jscatter as js calibration = js.sas.sasImage(js.examples.datapath+'/calibration.tiff') calibration.showPolar()
For offset detector
import jscatter as js import numpy as np sic = js.sas.sasImage(js.examples.datapath+'/Silicon.tiff') sic.setDetectorPosition([130,-100],0.070,90,90,0) sic.showPolar()
